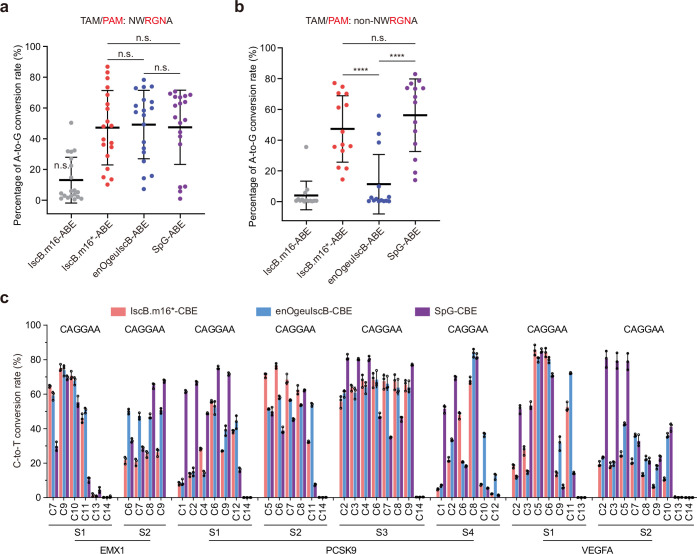
Extended Data Fig. 6. Base editing efficiency of IscB- and SpG-based adenine and cytosine base editors at endogenous loci in HEK293T cells.
a, b, Comparison of the A-to-G conversion efficiency of IscB.m16-ABE, IscB.m16*-ABE, enOgeuIscB-ABE, and SpG-ABE at 19 TAM sites of NWRGNA (a) and 14 TAM sites of NNNGNA, but non-NWRGNA (b). Data are collected from 19 and 14 endogenous sites, respectively, and presented as means ± s.d. Each dot represents the average highest base editing activity at each endogenous target site of three independent biological replicates. P values determined by Tukey’s multiple comparisons test following ordinary one-way analysis of variance. The Padj. are 0.613, 0.00005 and 0.0000007, respectively. c, Comparisons of C-to-T conversion efficiency of IscB.m16-CBE, IscB.m16*-CBE, enOgeuIscB-CBE, SpG-CBE at 8 target sites containing various TAMs in HEK293T cells. Value and error bars are presented as means ± s.d., n = 3 independent biological replicates.