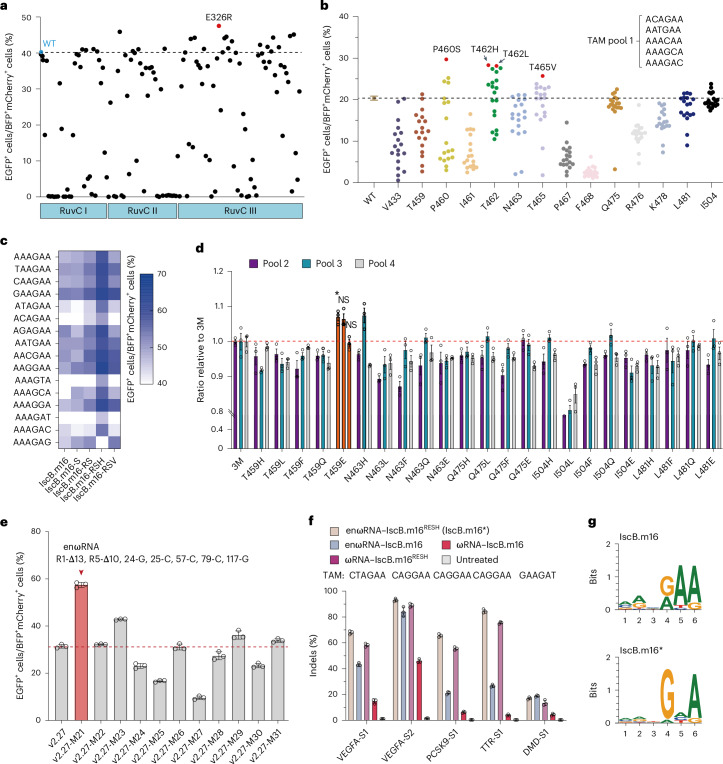
Fig. 3. Protein engineering of IscB.m16 to improve editing efficiency and expand TAM range in mammalian cells.
a, Screening for highly efficient variants by substitutions of amino acid residues in the RuvC domain of IscB.m16 protein with arginine. Each dot represents the editing activity for a single variant. The dashed line indicates the editing activity of the WT. b, Screening for highly efficient variants with saturation mutagenesis at selected sites using GFxxFP reporters containing a pooled TAM target. Each dot represents the editing activity for a single variant. The dashed line indicates the editing activity of the WT. c, Comparison of editing activity among WT IscB.m16 and its variants at 16 GFxxFP reporters with different TAMs. IscB.m16-S represents the variant P460S, IscB.m16-RS represents the variant with a combination of E326R and P460S, IscB.m16-RSH represents the variant with a combination of E326R, P460S and T462H and IscB.m16-RSV represents the variant with a combination of E326R, P460S and T465V. Colored dots reflect the mean of three independent biological replicates. d, Screening for variants with improved editing frequency based on GFxxFP reporters containing three different TAM pools along with the same ωRNA-v2.27. The orange bar represents the IscB.m16RESH variant with a combination of IscB.m16-RSH and T495E. P values were determined by Tukey’s multiple comparisons test following ordinary one-way ANOVA. *P < 0.05. NS, not significant. e, The second round of ωRNA engineering by substituting C•G base pairs on ωRNA-v2.27 (R1-Δ13, R5-Δ10, 24-G, 25-C, 57-G, 79-C and 117-C) based on IscB.m16RESH. f, Comparison of indel frequency of WT IscB.m16 and its variants at five endogenous sites in HEK293T cells. g, TAM logos of IscB.m16 and IscB.m16* systems. Values and error bars represent the mean ± s.d. (n = 3 independent biological replicates).