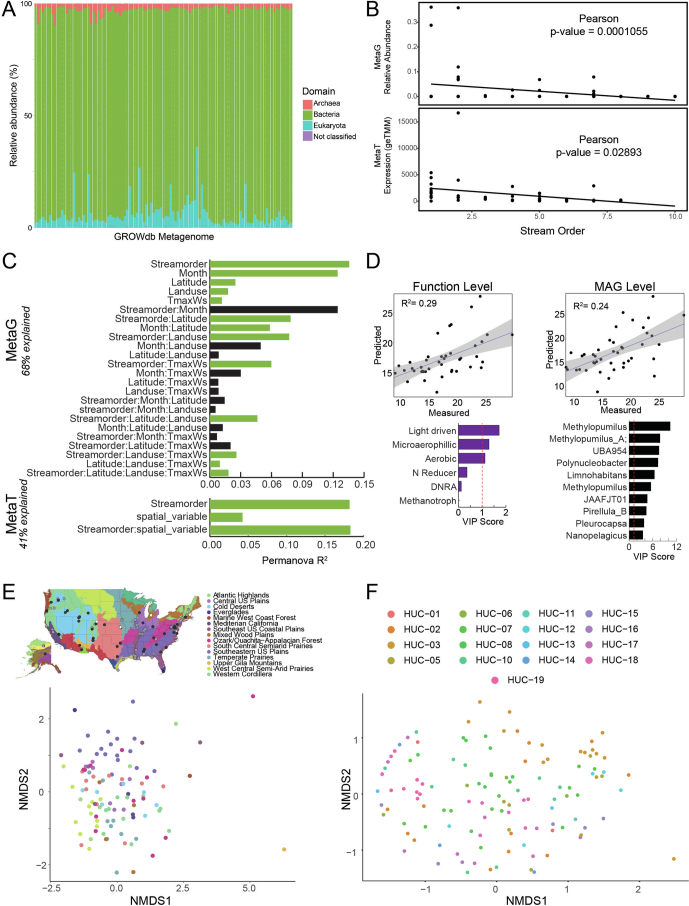
Extended Data Fig. 7. GROWdb membership and structure across geospatial parameters.
A) Stacked bar chart of the singleM profiles of GROWdb metagenomic reads, with bars coloured by domain. By domain, the most reads are assigned to the Bacteria (mean=91.1%), followed by Eukaryota (mean=6.1%), Archaea (mean=2.6%), and Unknown (mean=0.2%). B) Correlations of Patescibacteria relative abundance (metagenomics, top) and expression (metatranscriptomics, bottom) with stream order. Correlation significance was tested in R using cor.test (two-sided), with p-values shown. C) Permutational analysis of variance (PERMANOVA) results for metagenomes (metaG) and metatranscriptomes (metaT) indicate that drivers of community structure and expression, respectively. These drivers and their interactions explain 68% of the metagenome and 41% of the metatranscriptome variance. Bar height represents the R2, with green bars denoting significant drivers (p < 0.05), while black bars are not significant drivers. D) Sparse Partial Least Squares (sPLS) regressions show significant function (top) and MAG level (bottom) expression predictions of watershed maximum temperature, with key variables (Variable Importance Projection >1) denoted in bar graphs below. Fitted regression line is shown with grey shading representing 95% confidence interval. E) Non-metric multidimensional scaling of genome resolved metagenomic Bray-Curtis distances shows clustering of microbial communities by ecoregion (classified by Omernik II), with sampling location depicted on map above (mrpp, p < 0.001). Abbreviations: NPOC, Non-Purgable Organic Carbon; DNRA, Dissimilatory Nitrite Reduction to Ammonia; WWTP Density, Waste Water Treatment Plant Density; NPP, Net Primary Production. F) Non-metric multidimensional scaling of genome resolved metagenomic Bray-Curtis distances shows clustering of microbial communities by hydrological unit (HUC-2), with sampling location depicted on map on Fig. 1c.