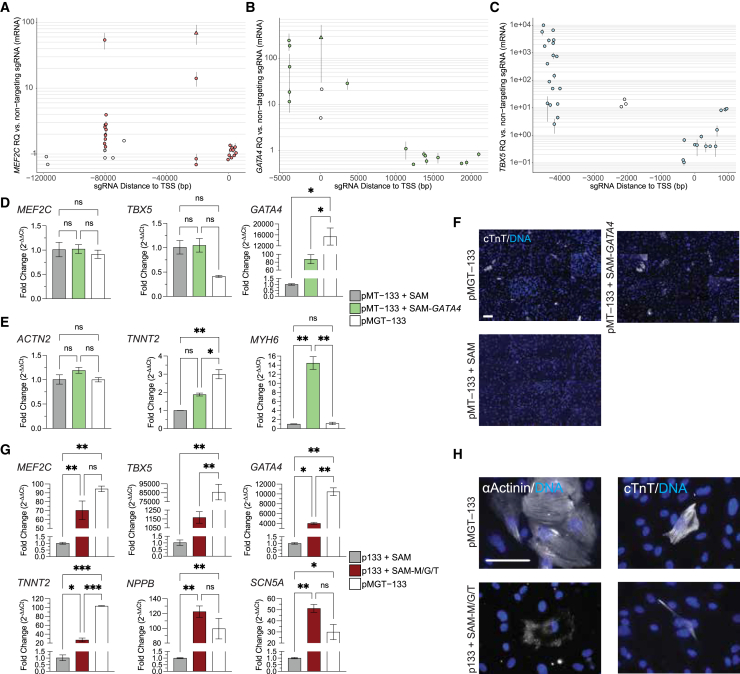
Figure 6.
Reprogramming H9Fs into iCMs via CRISPRa-mediated activation of GATA4, MEF2C, and TBX5
(A–C) Summary of sgRNAs tested for CRISPRa-mediated activation of MEF2C (A), GATA4 (B), and TBX5 (C). Data represent six independent experiments. Point range plots illustrate the activation levels induced by each sgRNA and their relative position to the TSS. Open circles, one replicate; filled circles, two replicates, one experiment; filled triangles, three or more replicates, two or more experiments. (D–F) Replacement of retroviral overexpressing GATA4 with SAM-GATA4. (D) qPCR analysis at day 2. The y axis indicates fold change by 2−ΔΔCt relative to control pMT−133+SAM. n = 2 biological replicates. (E) qPCR analysis at day 10. The y axis indicates fold change by 2−ΔΔCt relative to the pMT−133+SAM control. n = 2 biological replicates. (F) Tiled composite images of ICC staining for cTnT at day 14, with 40× images captured from 16 random fields per well. (G and H) CRISPRa activation of all three reprogramming factors with SAM-MGT (G) qPCR at day 14. The y axis indicates fold change by 2−ΔΔCt relative to the pMT−133+SAM control. n = 2 biological replicates. (H) Representative images of ICC staining for sarcomere components at day 14, acquired at 40× magnification. ns, not significant. Scale bars, 100 μm ∗p < 0.05; ∗∗p < 0.01.