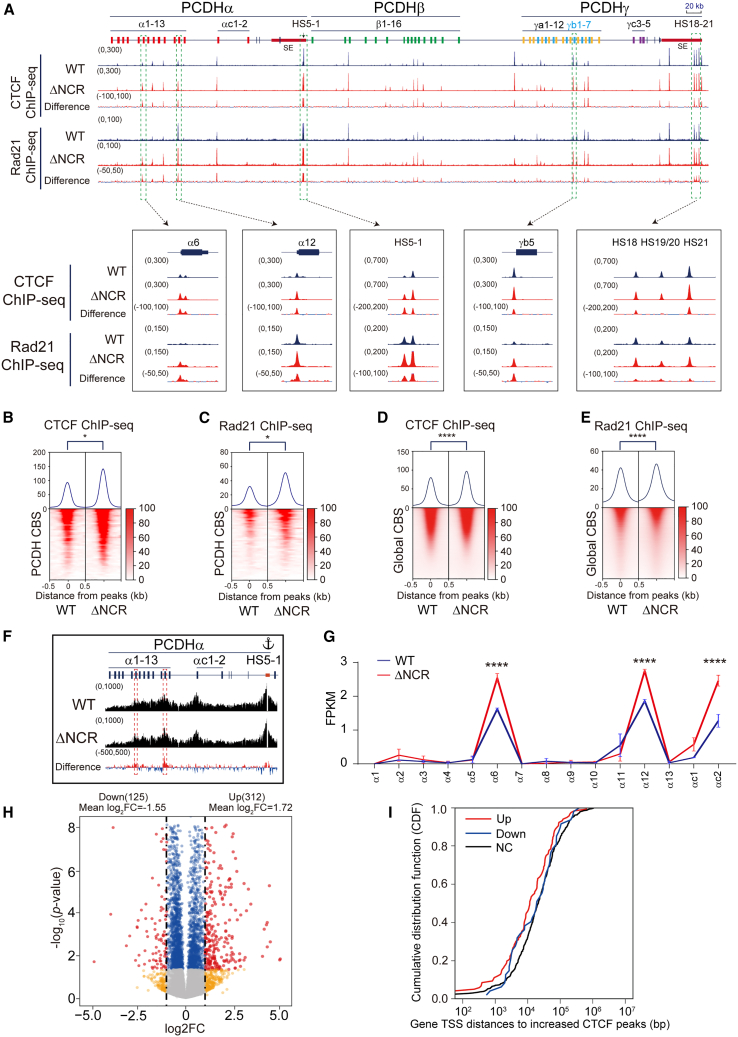
Figure 4.
NCR deletion increases CTCF binding and affects gene regulation
(A) CTCF and Rad21 ChIP-seq signals at the cPCDH locus, showing increased CTCF and cohesin enrichments at CBS elements upon NCR deletion (Δ612-637).
(B and C) Heatmaps of CTCF (B) and Rad21 (C) ChIP-seq signals at the cPCDH locus. Student’s t test, ∗p < 0.05.
(D and E) Heatmaps of CTCF (D) and Rad21 (E) ChIP-seq signals, indicating genome-wide CTCF and Rad21 enrichments upon NCR deletion. Student’s t test, ∗∗∗∗p < 0.0001.
(F) Quantitative high-resolution 4C (QHR-4C) experiments with HS5-1 as an anchor, indicating increased chromatin interactions of the HS5-1 enhancer with PCDHα6 or PCDHα12 promoters.
(G) RNA-seq indicates increased expression levels of PCDHα6, PCDHα12, and PCDHαc2 upon NCR deletion. Data are presented as mean ± SD; Student’s t test, ∗∗∗∗p < 0.0001.
(H) Volcano plots of differential gene expression analyses for WT and ΔNCR cells. Red dots, fold change of gene expression upon NCR deletion (log2FC > 1 and adjusted p value <0.05). Blue dots, genes only passed adjusted p value <0.05. Yellow dots, gene only passed log2FC > 1. FC, fold change.
(I) TSS distances of up-, down-, or none-regulated (NC) genes to the closest increased CTCF peaks in ΔNCR cells (data are shown as a cumulative distribution function, CDF).
See also Figures S1, S3, and S6 and Tables S5 and S6.