Figure 1.
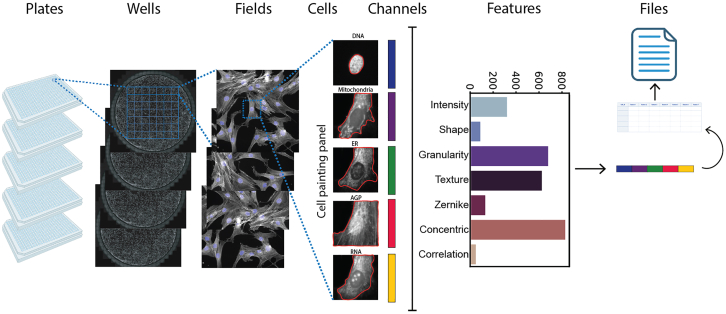
Overview of ScaleFEx’s pipeline on a Cell Painting assay
For each experiment, ScaleFEx queries the plate’s images and starts a serialized computation (parallelized over multiple machines per plate in the AWS version). Next, the wells are distributed and computed in parallel according to the number of workers specified by the user. Within each worker, after locating the centroids of the nuclei, each channel image is loaded into memory and cropped around the centroid. For each image a segmentation mask is computed, and starting from there all of the subsequent channel based features. Once the full feature vector is computed (containing measurements related to shape, texture, Zernike moments, granularity, intensity, concentric rings, colocalization, mitochondria and RNA), the vector is appended to the experiment’s csv file, to avoid needing to store the entire dataset into the random-access memory (RAM). See also Figure S1.