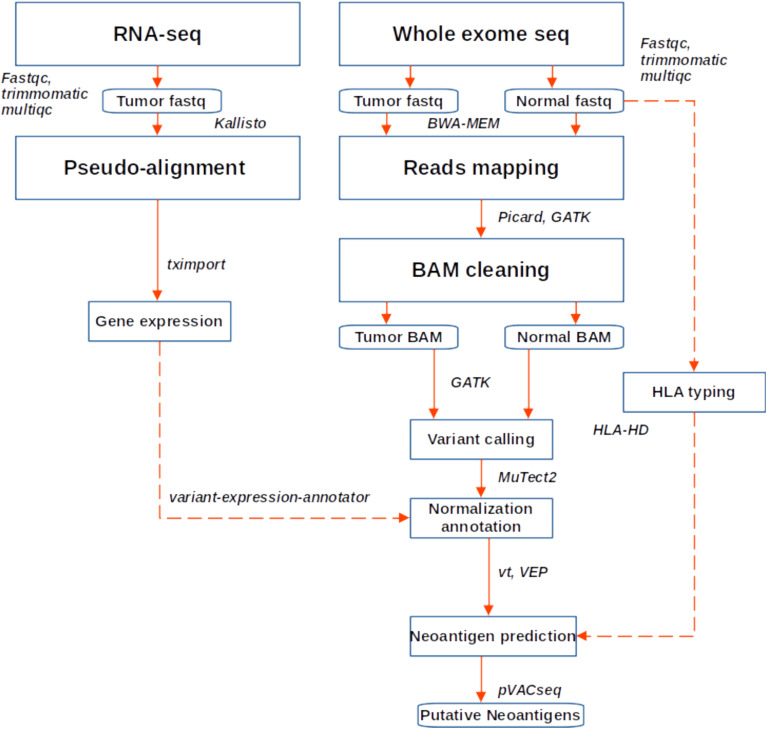
Figure 1.
Workflow for neoantigen prediction from WES and RNA sequencing data. Fastq files were quality checked, trimmed and aligned to the hg38 genome. Variant calling was performed following GATK best practice, while gene expression was quantified using Kallisto. Variants were annotated and expression data added, after which neoantigen prediction was performed in PVACseq pipeline.