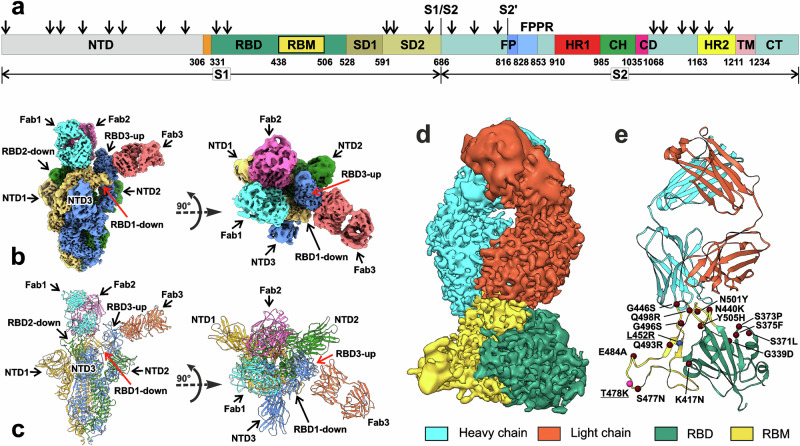
Fig. 1. Cryo-EM structure of the full-length Delta SARS-CoV-2 S-protein in the complex with REGN10987-Fab.
a Schematic representation of the sequence of full-length SARS-CoV-2 S-protein. Segments of S1 and S2 subunits include: NTD N-terminal domain, RBD receptor-binding domain, RBM receptor-binding module, SD1 and SD2, C-terminal subdomains 1 and 2; S1/S2, S1/S2 cleavage site; S2′, S2′ cleavage site; FP fusion peptide, FPPR fusion peptide proximal region, HR1 heptad repeat 1, CH central helix region, CD connector domain, HR2 heptad repeat 2, TM transmembrane helix, CT cytoplasmic tail; and arrow symbols for glycans. b, c Cryo-EM map of the Delta S-protein/Fab complex refined to 2.5 Å resolution and pseudoatomic model of the complex after local refinement of RBD/Fab regions. Three protomers (1, 2, and 3) of S-protein and the attached Fabs are color coded. RBD1/Fab1 and RBD2/Fab2 complexes are in ‘down’’ conformation, while RBD3/Fab3—in ‘up’’. d, e Cryo-EM map and modeled structure of the RBD1/Fab1 complex after focused refinement to 3.2 Å. The sites of Delta (blue and magenta spheres, underlined) and Omicron BA.1 (magenta and brown spheres) mutations are shown.