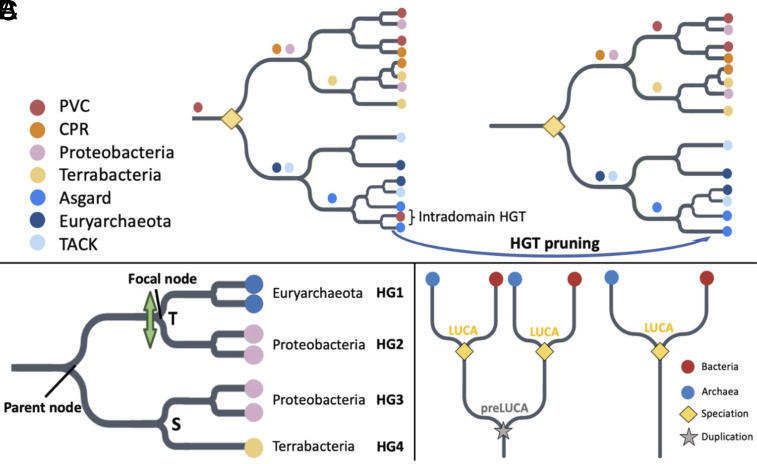
Fig. 2.
Criteria for (A) LUCA Pfam annotation, (B) identifying HGT to be filtered, and (C) pre-LUCA Pfam annotation. Details are in Methods, with a brief summary here. (A) Pruning HGT between archaea and bacteria reveals a LUCA node as dividing bacteria and archaea at the root. Colored circles are indicated just upstream of the most recent common ancestor (MRCA) of all copies of that Pfam found within the same taxonomic supergroup. We recognize a total of five bacterial supergroups [FCB, PVC, CPR, Terrabacteria, and Proteobacteria (25, 26)] and four archaeal supergroups [TACK, DPANN, Asgard, and Euryarchaeota (27, 28)]; only 4 out of 5 bacterial supergroups and 3 out of 4 archaeal supergroups are shown. The yellow diamond indicates LUCA as a speciation event between archaea and bacteria. We do not assume that the LUCA coalescence timing was the same for every Pfam (29). Prior to HGT pruning, PVC sequences can be found on either side of the two lineages divided by the root. After pruning intradomain HGT, four MRCAs are found one node away from the root, and three more MRCAs are found two nodes away from the root, fulfilling our other LUCA criterion described in the Methods, namely the presence of at least three bacterial and at least two archaeal supergroup MRCAs one to two nodes away from the root. (B) Criteria for pruning likely HGT between archaea and bacteria (see Materials and Methods for details). We partition into monophyletic groups of sequences in the same supergroup; in this example, there are four such groups, representing two bacterial supergroups and one archaeal supergroup. There is one “mixed” node, separating an archaeal group (HG1) from a bacterial group (HG2). It is also annotated by GeneRax (19) as a transfer “T.” The bacterial nature of groups 3 and 4 indicates a putative HGT direction from group 2 to group 1. Group 2 does not contain any Euryarchaeota sequences, meeting the third and final requirement for pruning of group 1. If neither Proteobacteria nor Euryarchaeota sequences were present among the other descendants of the parent node, both groups 1 and 2 would be considered acceptors of a transferred Pfam and would both be pruned from the tree. (C) Pre-LUCA Pfams have at least two nodes annotated as LUCA.