Fig. 3.
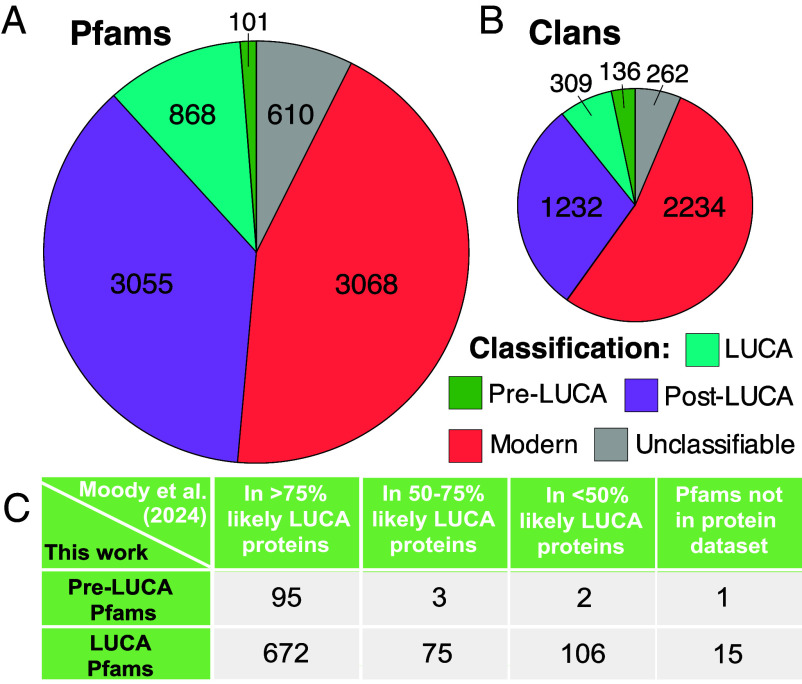
Pfams (A) and clans (B) classified as ancient are well validated by the whole gene annotations of Moody et al. (21) (C). (A) Ancient post-LUCA Pfam classifications include 285 LACA candidates and 2,770 LBCA candidates (more analysis would be required to rule out extensive HGT within archaea or bacteria). Modern Pfams are distributed among the prokaryotic supergroups as follows: 9 CPR, 210 FCB, 942 Proteobacteria, 51 PVC, 1,111 Terrabacteria, 2 Asgard, 49 TACK, and 177 Euryarchaeota. In addition to supergroup-specific modern Pfams, we classified another 1,097 Pfams, present in exactly two bacterial supergroups, as modern post-LBCA. We deemed 15 Pfams unclassifiable due to high inferred HGT rates, 397 due to uncertainty in rooting, and 198 due to ancient rooting combined with absence from too many supergroups (Materials and Methods). (B) Pre-LUCA clans contain at least two LUCA-classified Pfams or one pre-LUCA Pfam, whereas LUCA clans contain exactly one LUCA Pfam. Ancient post-LUCA clans contain no LUCA, pre-LUCA, or unclassified Pfams; they include an ancient post-LUCA Pfam or at least two modern Pfams covering at least two supergroups from only one of either bacteria or archaea. Modern clans include Pfams whose root is assigned at the origin of one supergroup. Finally, unclassifiable clans did not meet any of our clan classification criteria, e.g., because they included both post-LUCA and unclassifiable Pfams. (C) 98% of our pre-LUCA Pfams and 87% of our LUCA Pfams are present in genes annotated by as present in LUCA with more than 50% confidence, when present in their dataset. We mapped all Clusters of Orthologous Genes (COGs) (30) in Moody et al. (Supplementary Table 1 in ref. 21 to UniProt IDs (31) using the EggNOG 5.0 database (32). We then identified their associated Pfams using the “Pfam-A.regions.uniprot.tsv” file downloaded from the Pfam FTP site (https://pfam-docs.readthedocs.io/en/latest/ftp-site.html#current-release) (24) on May 28th, 2024. Our protein to Pfam ID mappings are available in “Protein2Domain_mappings” in ref. 33.