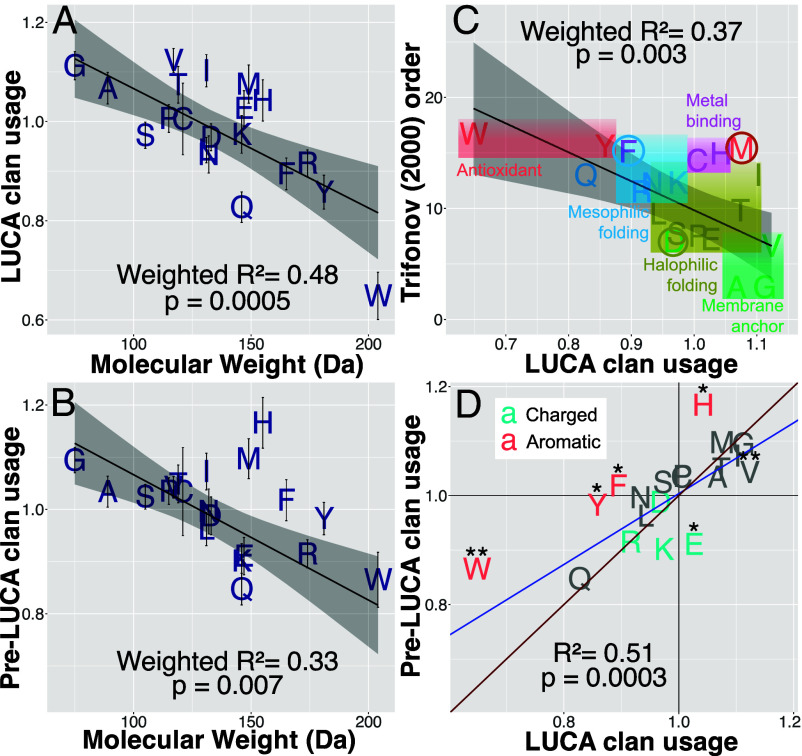
Fig. 4.
LUCA is enriched for smaller amino acids, with subtle differences between single-copy LUCA vs. multicopy pre-LUCA sequences. Ancestrally reconstructed amino acid frequencies in LUCA and pre-LUCA clans are shown relative to those in ancient post-LUCA clans. (A) LUCA clans and (B) pre-LUCA clans are enriched for amino acids of smaller molecular weight. Weighted model 1 regression lines are shown in black with 95% CI gray shading. Error bars indicate SE. (C) Character colors show the assignments of Moosmann (5); colored circles indicate our reassignments. We reclassify F because phenylalanine is enriched in proteins in mesophiles compared to their orthologs in thermophiles and hyperthermophiles (47). We reclassify D because the surfaces of proteins within halophilic bacteria are highly enriched in aspartic acid compared to in the surfaces of nonhalophilic mesophilic and thermophilic bacteria, in a manner that cannot be accounted for by the dinucleotide composition of the halophilic genomes (48). The brown circle around M highlights that while methionine might not be utilized against reactive oxygen species, it might once have been against ancient reactive sulfur species. (D) Model 2 Deming regression [accounting for SE in both variables, implemented in deming() version 1.4-1 (49) in blue shows that pre-LUCA enrichments are not more extreme versions of LUCA enrichments, lying on the wrong side of the y = x red line. We include the imidazole-ring-containing H as aromatic. Asterisks (*) indicate statistically different amino acid frequencies between pre-LUCA and LUCA (Welch two-sample t test, P < 0.05 and P < 0.01).