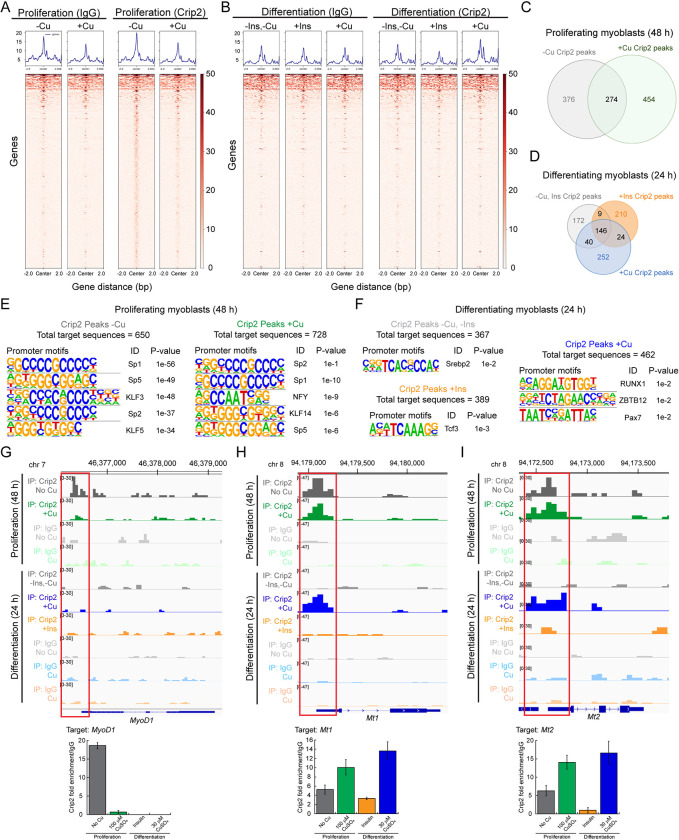
Fig 5. mCrip2 binding to chromatin in proliferating and differentiating myoblasts.
Representative heat maps from peak calling by Sparse Enrichment Analysis (SEACR) for CUT&RUN. IgG controls and mCrip2 binding are depicted. A. Proliferating WT primary myoblasts were supplemented or not with 100 μM CuSO4. B. Differentiating (24 h) WT primary myoblasts were supplemented or not with insulin of 30 μM CuSO4. Overlap of CUT&RUN peaks of mCrip2 across the genome observed in proliferating (C) and differentiating (D) cells in the presence or absence of Cu and insulin. See complete set of genes in S4 Table. Main changes of mCrip2 motif-binding dependent on Cu supplementation in proliferating and differentiating primary myoblasts. Novel consensus DNA-binding motifs identified from peak calling by Sparse Enrichment Analysis (SEACR) for CUT&RUN within mCrip2 peaks in proliferating cells (E) and differentiating myoblasts (F) supplemented or not with Cu. The top five most significant motifs enriched, including the DNA logo, its corresponding TF, and its P value are shown. See S4 Table for complete list of peaks. Representative genome browser tracks of CUT&RUN experiments examining mCrip2 binding to the MyoD1 (G), Mt1 (H) and Mt2 (I) promoters in proliferating and differentiating myoblasts (upper panels). MyoD1, M1 and Mt2 promoters were selected as a representative locus for validation by ChIP-qPCR (lower panels). Plots represent data obtained from 5 independent biological experiments.