Figure 1.
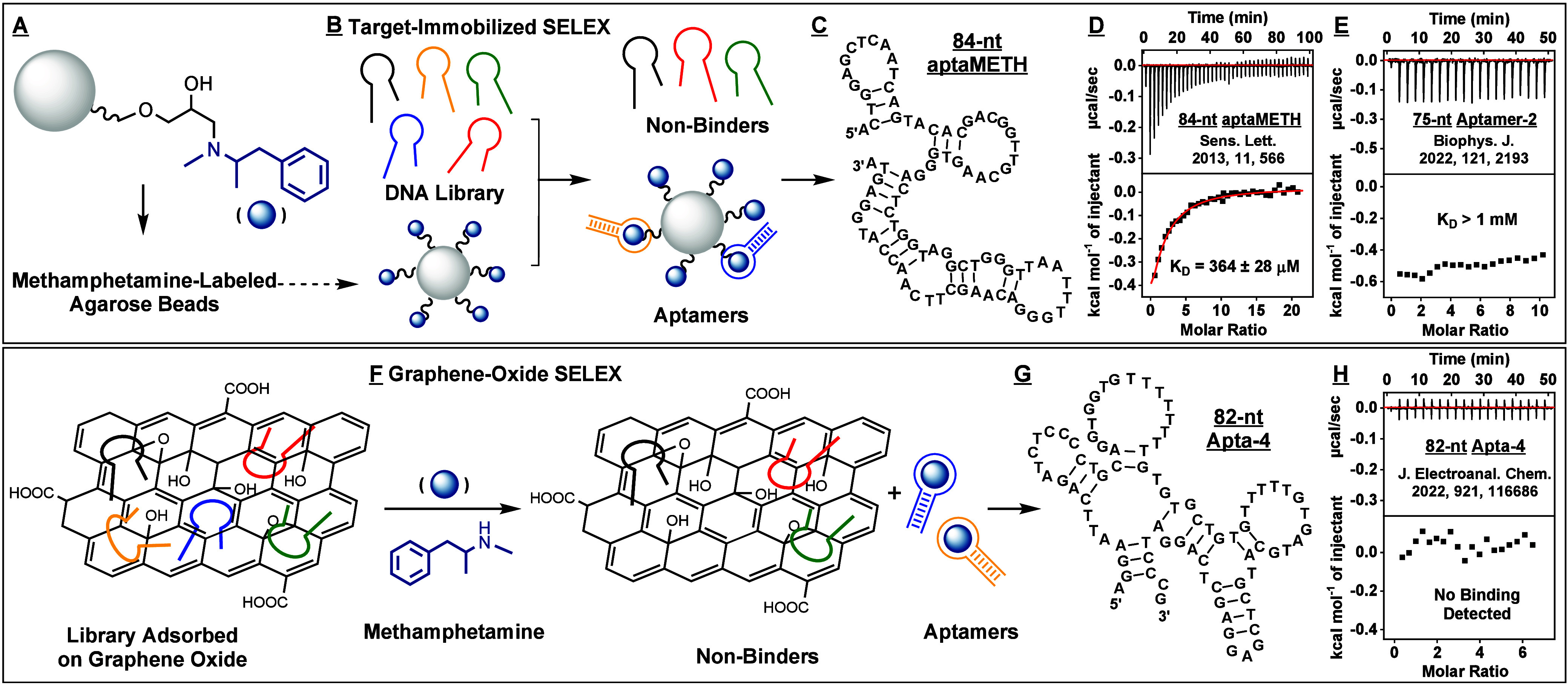
Affinity characterization of aptamers previously isolated for methamphetamine by other groups. Aptamers have been previously isolated using target-immobilized SELEX, in which (A) methamphetamine is conjugated to agarose beads via its amino group. (B) These beads are incubated with the DNA library, and binders are physically partitioned from binding-incompetent sequences. (C) Ebrahimi et al.22 identified 84-nt aptaMETH in this way and reported that this aptamer binds (+)-methamphetamine with a KD of 100 nM. (D) However, our isothermal calorimetry (ITC) results showed a far higher KD of 364 μM in their selection buffer. (E) Sester et al.24 used a similar approach to isolate 75-nt Aptamer-2, with a reported KD of 244 nM, but our ITC results again indicated a higher KD of >1 mM for (+)-methamphetamine in their reported selection buffer. (F) Bor et al.23 utilized graphene-oxide SELEX to isolate DNA aptamers for methamphetamine, based on binding-induced desorption of target-specific aptamers from graphene oxide. (G) The resulting 82-nt Apta-4 aptamer reportedly bound methamphetamine with a KD of 1.3 μM. (H) In contrast, our ITC results indicate no binding at all in their reported selection buffer.
