Figure 2.
HHRzs in Arabidopsis.
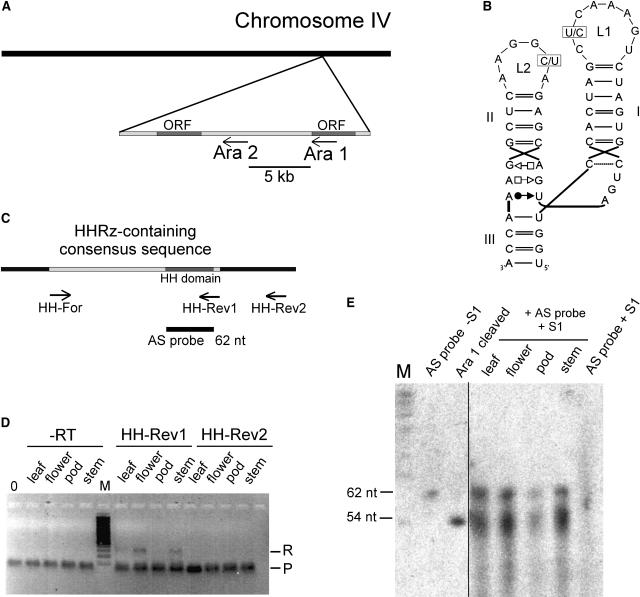
(A) Chromosomal localization of the two motifs in the genome of Arabidopsis. The HHRz sequences Ara1 and Ara2 are found within 5 kb in the antisense orientation to open reading frames (ORFs) on chromosome IV.
(B) Secondary structures of the HHRz domains. Variable positions in loops L1 and L2 are indicated. In Ara1, the variable positions are occupied by a C in loop L1 and a U in loop L2, whereas in Ara2, these nucleotides are replaced by U (loop L1) and C (loop L2).
(C) Design of primers and antisense RNA used in RT-PCR and the S1 nuclease assay. The HHRz-containing 300-nucleotide consensus sequence is shown in light gray, and the 66-nucleotide hammerhead motif within is shown in dark gray. The positions of primers HH-Rev1, HH-Rev2, and HH-For for RT-PCR are indicated, as is the position of the 62-nucleotide (nt) antisense (AS) probe used in the S1 nuclease assay.
(D) RT-PCR of RNA isolated from the indicated organs. For first-strand cDNA synthesis, the specific primers HH-Rev1 and HH-Rev2 were used. PCR was performed using the specific primers HH-Rev1 and HH-For that amplify the uncleaved 300-nucleotide HHRz-containing consensus sequence. R denotes the signal of the RT-PCR product, and P indicates a band generated by the primers. In the control reactions (−RT), PCR was performed on RNA that had not been subjected to first-strand cDNA synthesis, and 0 refers to a template-free control PCR. M denotes a 100-bp marker. Bands were visualized by ethidium bromide staining.
(E) Nuclease S1 protection assay. Total RNA (0.5 μg) of the indicated tissues was hybridized to a 62-nucleotide antisense RNA of the hammerhead domain. S1 nuclease was used to digest single-stranded RNA. As size markers, a non-S1–treated antisense probe and an in vitro transcript of the 66-nucleotide Ara1 sequence, which self-cleaves in the course of the transcription, were included, resulting in fragments of 54 and 12 nucleotides (the latter not visible on the gel). As a negative control, the antisense RNA probe was digested with nuclease S1 (AS probe + S1). RNA was visualized by exposure to x-ray films.