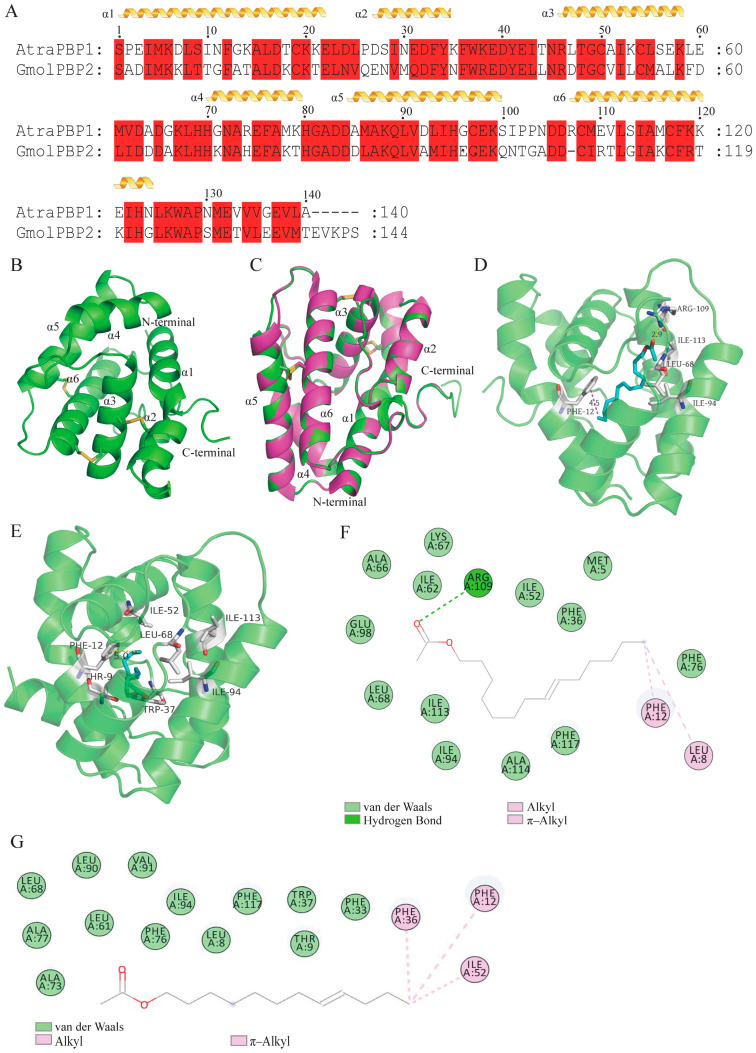
Figure 4.
Construction of a 3D model of GmolPBP2 and analysis of the amino acid residues and interaction forces in GmolPBP2–Z8-14:Ac and GmolPBP2–Z8-12:Ac. (A) Sequence alignment of GmolPBP2 and AtraPBP1. α-helices are displayed as squiggles, while residues that are strictly identical are highlighted with a red background. (B) Predicted 3D model of GmolPBP2. (C) Structure alignment of AtraPBP1 (4INW) and GmolPBP2. (D,E) represent the predicted amino acid residues and interaction forces of GmolPBP2 binding to Z8-14:Ac and Z8-12:Ac, respectively (3D structure). (F,G) represent the predicted amino acid residues and interaction forces of GmolPBP2 binding to Z8-14:Ac and Z8-12:Ac, respectively (2D structure).