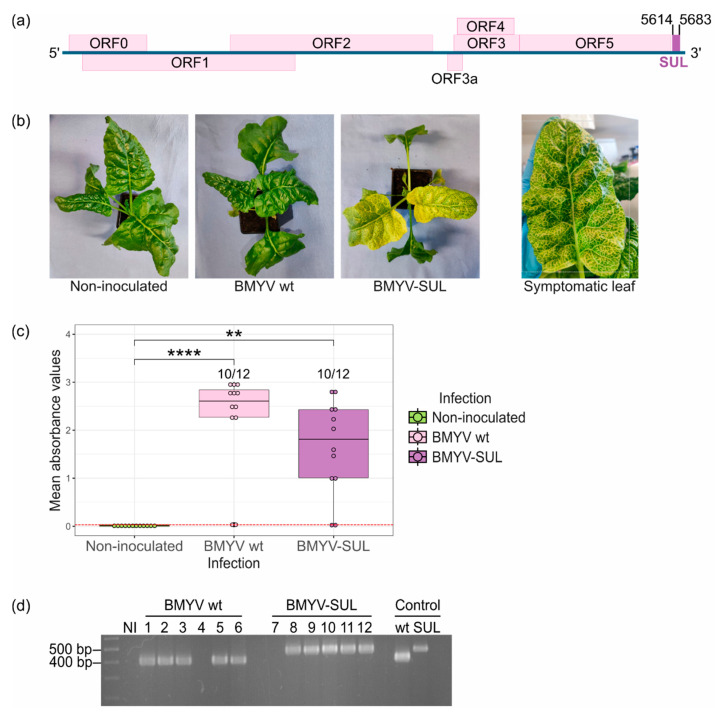
Figure 1.
Infectivity assay of agrobacterium BMYV-SUL clone in B. vulgaris plants (line A). (a) Schematic representation of BMYV-based VIGS vector construction. A 70 nts long fragment of the endogenous B. vulgaris SUL gene, shown as a purple rectangle, was inserted at position 5614 of the viral genome. (b) Symptoms of vein clearing and leaf yellowing observed on sugar beets agroinoculated with the recombinant BMYV-SUL clone three weeks post-infiltration. No symptoms were observed on non-agroinoculated plants or plants which were agroinoculated with the wild-type (wt) BMYV clone. (c) Virus accumulation analysed by TAS-ELISA. Each dot of the boxplots represents the absorbance value recorded for each plant. Values from non-inoculated samples are shown in green, of wild-type BMYV agroinoculated plants in pink, and of recombinant BMYV-SUL in purple. The red dotted line indicates the positivity threshold. Statistical analyses were performed using a Kruskal–Wallis test followed by a Dunn’s test for pairwise comparisons and Bonferroni correction. The stars above the bars indicate the statistically significant differences in absorbance values for the three infection conditions. p-values less than 0.01 and 0.0001 are respectively flagged (**) and (****). (d) Stability of the SUL insert verified by RT-PCR. The first lane corresponds to the 10 kb MassRulerTM DNA ladder, ‘NI’ refers to non-agroinoculated samples, lanes 1 to 6 correspond to the wild-type BMYV agroinoculated samples, and lanes 7 to 12 correspond to recombinant BMYV-SUL samples. Plasmid DNA of the wild-type and recombinant BMYV constructs were used as PCR size controls, as shown in the last two lanes.