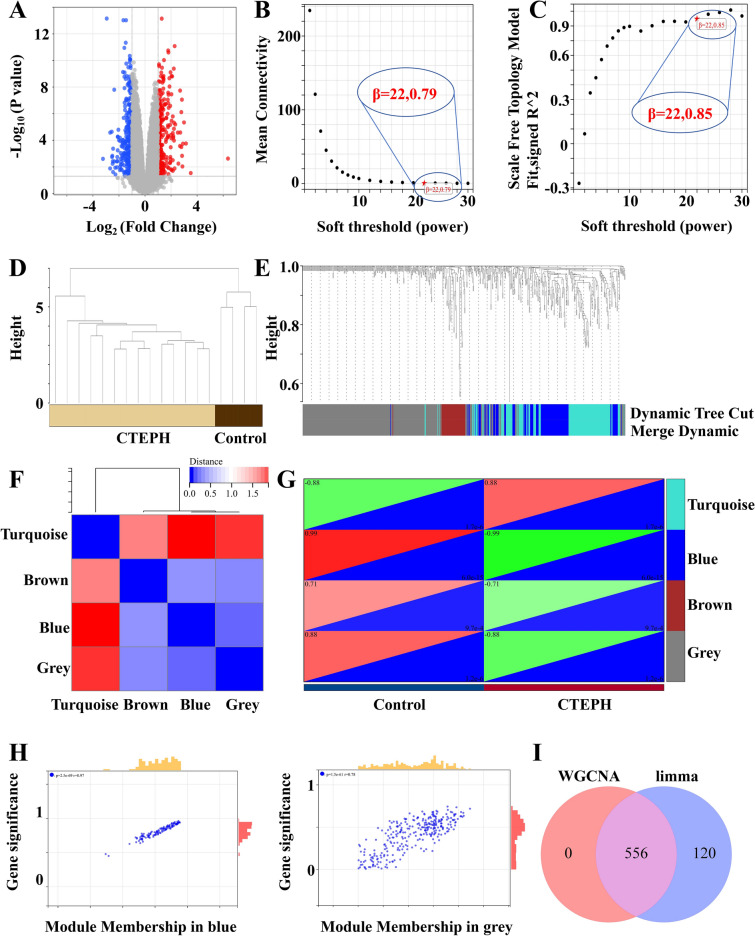
Fig. 2.
Identification of differentially expressed genes in CTEPH by limma and WGCNA packages. (A) PCA diagram of the DEGs expression. Red represents gene up-regulation and blue represents gene down-regulation. (B) Scale independence diagram. (C) Average connectivity graph. The soft threshold is 22. (D) Hierarchical clustering tree of MRNA expression patterns of 18 samples. Sample aggregation, no outlier samples. (E) Correlation diagram between modules. (F) Dendrogram of all differentially expressed genes clustered based on different similarity measures. Turquoise, gray, blue and brown are the four modules with the highest degree of clustering. (G) Heat map of the characteristic genes of the module. (H) Scatterplot of correlation between modules and clinical traits. Gray (r = 0.78, p = 1.5e−61), blue (r = 0.63, p = 2.7e−17). (I) Intersection gene map of limma package and WGCNA package.