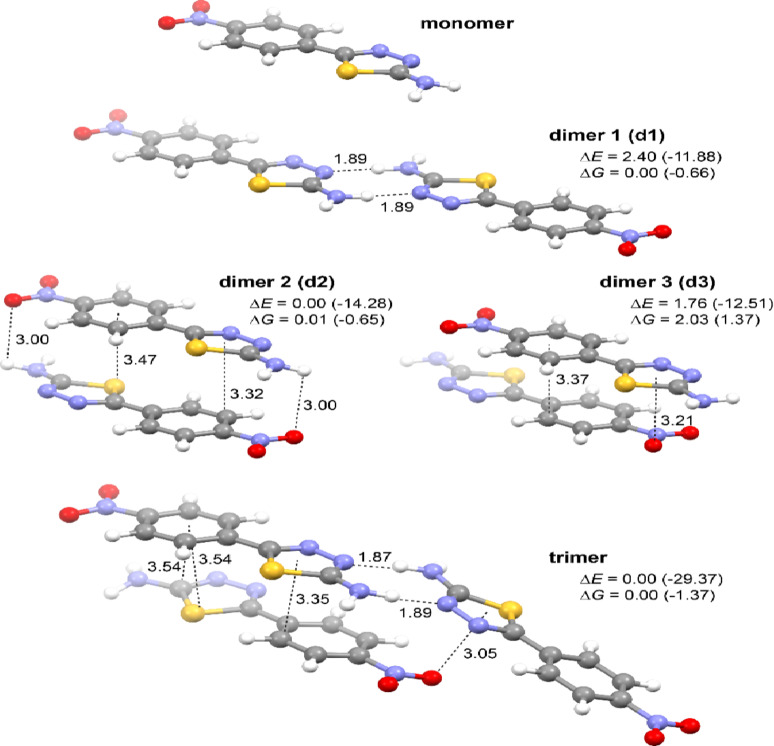
Fig. 10.
Optimized (DFT-B3LYP + D3/6‑31 + + G(d, p)/PCM(PrOH)) molecular structures of the monomer and aggregated models (dimers and trimer) of NTA. Values listed are hydrogen bonding and π‧‧‧ π stacking (measured between the ring centroid and the closest atom in the neighboring molecule) distances (in Å) and relative energy ΔE and free energy ΔG values (in kcal/mol). In parentheses, the corresponding formation energy and free energy values are listed, computed as a difference between the energy of the aggregate and the sum of the energy of its components that is the monomer in its optimal geometry. For the structures of the aggregated models of NTA in their S1 excited state, see Figure S2.5.