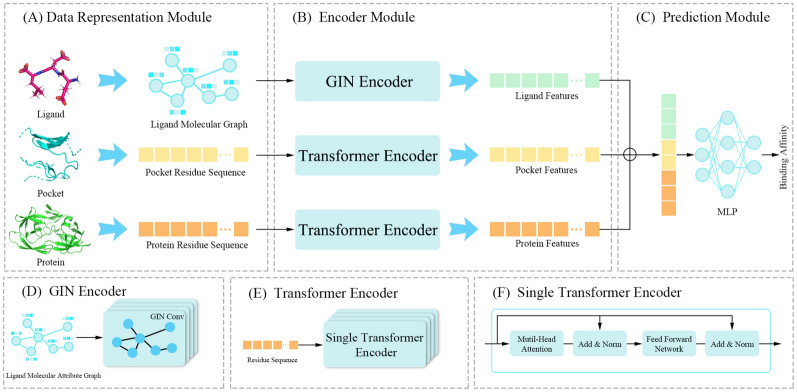
Fig. 2.
Architecture of DeepTGIN. A Data representation module: This module includes three inputs: the ligand molecular graph, the pocket residue sequence, and the protein residue sequence. B Encoder Module: This module learns the features of the ligand molecular graph using the GIN Encoder and learns the features of the protein residue sequence and pocket residue sequence using the Transformer Encoder. C Prediction Module: This module predicts the binding affinity. D GIN Encoder: The GIN Encoder comprises 4 layers of GIN. E Transformer Encoder: The Transformer Encoder consists of 4 layers of a single transformer encoder. F Single Transformer Encoder: Details about the single transformer encoder