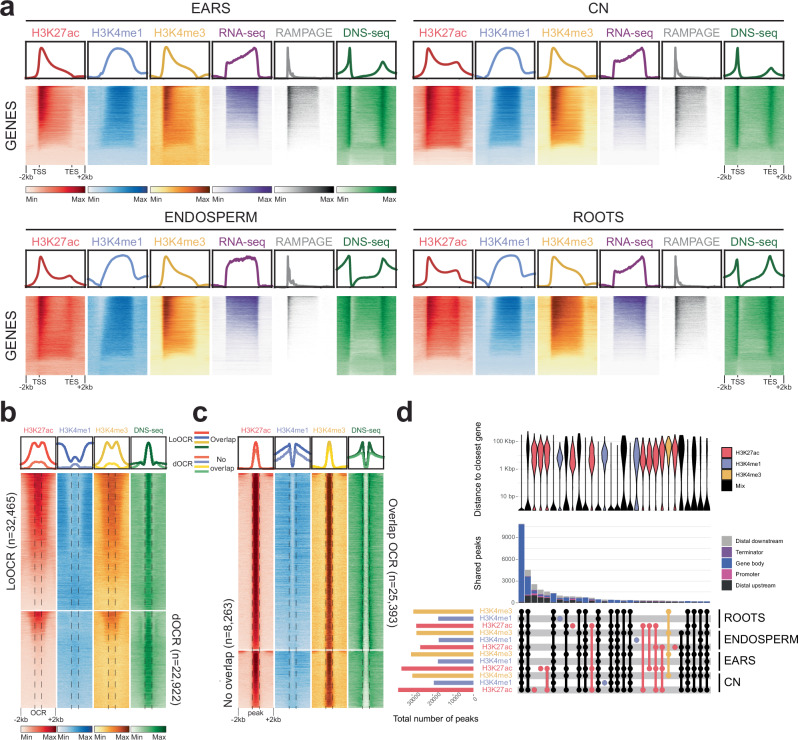
Fig. 1. Histone H3 modifications mark DNA regulatory elements in maize inbred lines.
a Heatmaps and metaplots of H3K27ac, H3K4me1, H3K4me3, RNA-seq, RAMPAGE and differential nucleosome sensitivity (DNS-seq)35 over all annotated genes in each tissue of B73 (NAM reference genome), scaled to the same size, with 2 kb upstream and downstream. CN = coleoptilar node. b Heatmaps and metaplots of B73 ears H3K27ac, H3K4me1, H3K4me3 and DNS-seq in local and distal open chromatin regions (LoOCR and dOCR, respectively) previously identified by ATAC-seq38. Bona fide regulatory elements are enriched for H3K27Ac and H3K4me3 but not H3K4me1. c Heatmaps and metaplots of H3K27ac, H3K4me1, H3K4me3 and DNS-seq at all H3K27ac peaks (regulatory elements) in B73 ears. 25,393 peaks intersect previously identified OCRs (20,334 LoOCRs and 5059 dOCRs) but 8263 peaks do not overlap. d Summary of shared ChIP-seq peaks in W22 (v2 reference genome). The Upset plot (lower panel) displays the overlap between H3K27ac, H3K4me1 and H3K4me3 peaks in the four tissues. The total number of peaks in each sample is shown on the histogram on the left-hand side of the intersection matrix, while the number of shared peaks between samples is shown above (middle panel), color coded by genomic feature. The violin plot (upper panel) compares the distance between peaks and the closest gene. Tissue specific peaks are mostly at distal elements, whereas loci with several histone marks in multiple tissues are mostly at annotated genes. Distal regulatory elements lie between 2 kb and 100 kb from the nearest gene.