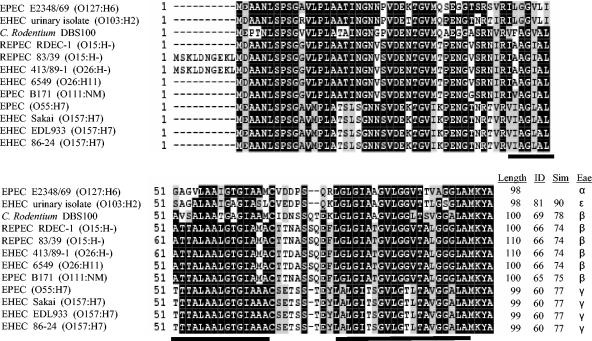
FIG. 1.
Alignment of SepZ from EPEC strain E2348/69 with SepZ from an EHEC strain isolated from the urine of a patient with hemolytic uremic syndrome, murine pathogen Citrobacter rodentium strain DBS100, rabbit pathogen REPEC strain RDEC-1, rabbit pathogen REPEC strain 83/39, EHEC strain 413/89-1, EHEC strain 6549, EPEC strain B171, an EPEC strain of serotype O55:H7, EHEC Sakai strain, EHEC strain EDL933, and EHEC strain 86-24 (accession numbers and references can be found in Materials and Methods). Numbers at the beginning of lines indicate amino acid positions. Black shading indicates identical residues shared by at least 50% of the sequences, while gray shading indicates similar but not identical residues shared by at least 50% of the sequences. Two predicted transmembrane helices are underlined. Dashes (-) indicate gaps or missing amino acids. The deduced amino acid sequences of REPEC 83/39 and EHEC 413/89-1 putatively contain an additional 10 amino acids at the N terminus. C. rodentium DBS100, RDEC-1, REPEC 83/39, EHEC 413/89-1, EHEC 6549, and EPEC B171 contain an additional two amino acids in the region corresponding to a putative loop between the two predicted transmembrane domains, while the EPEC serotype O55:H7 strain and the three EHEC serotype O157:H7 strains contain one additional amino acid in this region. Total lengths of the deduced sequences are listed at the end of the alignment, as are the percent identity (ID) and similarity (Sim) of the sequences in comparison to SepZ from EPEC E2348/69, and the corresponding intimin (eae) type (Eae) present in each strain.