Abstract
Phage-encoded murein hydrolases are either part of the lysis cassette or found as structural components of the phage virion. Here, we show that Staphylococcus aureus bacteriophage P68 contains a virion-associated muralytic enzyme. Protein 17 has a composite structure. The N-terminal part comprises the muralytic activity, whereas the C-terminal part is required for binding to the cell surface. A high multiplicity of infection with phage P68 caused rapid lysis, and purified protein 17 triggered premature lysis when added to S. aureus cells prior to infection with P68, suggesting that it functions to weaken the murein at the site of phage DNA entry. Protein 17 displayed activity against clinical S. aureus isolates, which are resistant to infection by phage P68, demonstrating that the protein targets surface structures distinct from the phage receptor. This broad activity spectrum of protein 17 could qualify virion-associated muralytic enzymes as attractive antimicrobials.
In 1940, Max Delbrück described two different types of phage-induced lysis (4) termed lysis from within (LI) and lysis from without (LO). LI occurs at a genetically determined time after infection. LI permits the release of phage progeny and generally depends on the activity of phage-encoded lysis genes (46). Double-stranded DNA phage, like phage λ, typically employ a holin-endolysin system to release phage progeny (46, 47). Endolysins form a group of unrelated enzymes with different muralytic activities that generally accumulate fully folded in the cytoplasm. Holins are small proteins, which form lesions in the cytoplasmic membrane through which phage-encoded endolysins gain access to the peptidoglycan at the end of the replication cycle. In contrast, small RNA or DNA phage encode neither an endolysin nor a holin. Phage progeny release seems to be achieved by phage-encoded proteins, which perturb the activity of host enzymes involved in peptidoglycan biosynthesis (3, 48).
LO was first described upon superinfection with phage T6 (4). Likewise, T4 virions caused rapid lysis of the host cell upon infection with high multiplicities of infection (MOI) (4). Phage T4 encodes two different lysozyme activities. The T4 gene e product is made during late protein synthesis and is responsible for degradation of the peptidoglycan layer of the host cell envelope during LI (35). Protein 5, with N-acetylmuramidase activity (13, 36), is anchored in the base plate structure of the virion tail (8, 14) and is responsible for LO. Similarly, protein P5, a lytic endopeptidase of the double-stranded RNA bacteriophage φ6 is associated with the nucleocapsid (30). More recently, the presence of peptidoglycan hydrolase activity has been demonstrated in the virions of Staphylococcus aureus phage φ11 and φ85 (32) as well as in the Lactococcus lactis phage Tuc2009 (16). In phage T7, the putative transglycosylase protein 16 was shown to increase the efficiency of phage genome translocation into the host cell (31, 33). In addition, homology searches have identified putative lytic transglycosylases among the structural proteins of phage PRD1 and P1 (18). PRD1 virions have been shown to contain two activities, the lysins P15 and P7. Neither protein is essential for infectivity, but P7 plays an accessory role in genome penetration into infected cells (38, 39).
In this study, we have identified protein 17 of the S. aureus lytic phage P68 (45) as a structural component of the virion with muralytic activity. The catalytic site was localized in the N-terminal part of the protein, whereas the C-terminal part represents the substrate recognition and/or binding domain. Protein 17 is the first virion-associated component characterized from a representative of the Podoviridae infecting S. aureus with broad activity against clinical S. aureus strains.
MATERIALS AND METHODS
Bacterial strains, phage, plasmids, and growth conditions.
Bacterial strains, phage, and plasmids used in this study are listed in Table 1. Staphylococcus aureus strains were grown in BHI broth (Merck) or on agar at 37°C. Escherichia coli strains were cultivated in Luria-Bertani (LB) broth at 37°C, and when appropriate, ampicillin (100 μg/ml) was added. Bacteriophage P68 was propagated on its S. aureus host 68 using double-layer agar plates, and progeny viruses were purified using CsCl density gradient centrifugation (40). P68 DNA was extracted and purified as previously described (45). Plaque formation of P68 on different S. aureus strains was assayed by following standard procedures (40).
TABLE 1.
Bacterial strains, plasmids, phage, and oligonucleotides used in this study
| Strain, plasmid, phage, or oligonucleotide | Genotype, description, or sequence (5′-3′)a | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP 10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 (ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3), carries T7 RNA polymerase gene 1 on λDE3 integrated into the chromosome | Invitrogen |
| S. aureus | ||
| 68 | Host for phage P68 | 45 |
| P1-P35 | Clinical isolates isolated from patients at the General Hospital in Vienna | This work |
| Phage P68 | 45 | |
| Plasmids | ||
| pet-16b | E. coli expression vector, Ampr | Novagen |
| pMUH17 | pET-16b + gene 17 of P68 | This work |
| pMUH17S | pET-16b + 1,440 bp of 5′ end of gene 17 of P68 | This work |
| pMUH17L | pET-16b + 1,782 bp of 5′ end of gene 17 of P68 | This work |
| Primers | ||
| I27 | TTTTTTAAACATATGGCATATAATGAAAACGATTTTAAATATTTTGATGACATT | |
| J27 | TTTTTTCTCGAGCTATTTTTGATGTTTTGCTACCCAACCATATTCACG | |
| K27 | CCCCCCCTCGAGCTATGCAGTACCCCCGTCAATATAATAAAAACCAGC | |
| L27 | CCCCCCCTCGAGCTAAATTTCTTCTGGCGCGTCTCTAAATAATTCCAT |
NdeI and XhoI sites are underlined.
DNA manipulations and plasmid construction.
Plasmid DNA was prepared with the QIAGEN midi plasmid kit. PCR amplifications were carried out using Pwo polymerase (Peqlab) and the primers listed in Table 1. The PCR fragments were purified using QIAquick spin PCR purification columns (QIAGEN). To assign the enzymatically active domain of protein 17, two 3′-truncated fragments of P68 gene 17 were generated by PCR.
The full-length N-terminally His6-tagged protein 17 (653 amino acids [aa]) was obtained as follows. The oligonucleotides I27 and J27 (Table 1) were annealed to template P68 DNA. The resulting PCR product was cleaved with NdeI/XhoI and cloned into plasmid pET-16b restricted with NdeI/XhoI (Table 1). This created plasmid pMUH17, which harbors an in-frame fusion between His6 codons and the entire gene 17.
Amplification of gene 17 with oligonucleotides I27 and K27 (Table 1) generated a 1,440-bp PCR fragment containing a 3′-terminally deleted gene 17 (codons 1 to 480) flanked by the restriction sites NdeI and XhoI. The NdeI/XhoI-restricted PCR fragment was cloned into pET-16b, resulting in plasmid pMUH17S. Likewise, the oligonucleotide pair I27-L27 (Table 1) was utilized to create a gene 17 fragment comprising codons 1 to 594. The 1,782-bp fragment was cloned into pET-16b, resulting in plasmid pMUH17L. All DNA cloning steps were initially performed with E. coli TOP 10.
Testing of S. aureus clinical isolates for methicillin resistance.
S. aureus clinical isolates were tested for methicillin resistance by the disk diffusion method using oxacillin as the class drug. Disk diffusion testing was performed according to the CLSI (formerly NCCLS) guidelines (37) using 1-μg-oxacillin-containing disks (Sigma-Aldrich) on Mueller-Hinton agar (Oxoid, England). The plates were incubated in ambient air at 35°C and read after 24 h. Interpretive zone diameters for oxacillin were as follows: ≤10 mm was resistant and ≥13 mm was susceptible. Intermediate zone sizes were between 10 and 13 mm. Any growth, including light growth within the 10-mm-diameter zone around the disk was taken to indicate resistance.
MALDI mass spectrometry.
Matrix-assisted laser desorption ionization (MALDI) mass spectrometry was carried out on a Bruker Reflex III time of flight mass spectrometer equipped with a 26-sample SCOUT source and video system, a nitrogen UV laser (λmax = 336 nm), and dual-channel plate detector (Bruker Daltonik, Bremen, Germany) using standard procedures.
Purification of protein 17 and variants thereof.
High-level synthesis of the His-tagged proteins 17 (653 aa), 17S (486 aa; aa 1 to 480 of protein 17), and 17L (600 aa; aa 1 to 594 of protein 17) was achieved in E. coli BL21(DE3) harboring plasmids pMUH17, pMUH17S, and pMUH17L, respectively. After overnight growth in Luria-Bertani broth at 37°C, the respective cultures were diluted 20-fold with LB medium and incubated at 37°C for 4 h when expression of the cloned genes was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After further incubation for 3 h at 37°C, the cells were harvested by centrifugation in a Sorvall RC 5C centrifuge at 6,000 rpm for 20 min. The cell pellets were resuspended in 10 ml lysis buffer (300 mM NaCl, 10 mM imidazole, 50 mM NaH2PO4, pH 8.0), and the cells were lysed by sonication (Branson Sonifier 250). The cell lysate was centrifuged at 28,000 × g for 30 min, followed by loading of the supernatant onto a 5-ml Ni-nitrilotriacetate-agarose (QIAGEN) column at 4°C. The N-terminal His-tagged proteins were eluted from the column with 250 mM imidazole followed by dialysis in 25 mM Tris (pH 7.5).
SDS-polyacrylamide gel electrophoresis and zymogram assays.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis was performed as described by Laemmli (19), and the zymogram assays were carried out as outlined by Lepeuple et al. (20). Briefly, 0.2% Staphylococcus aureus 68 autoclaved cells were included in 10% polyacrylamide gels for detection of bacteriolytic activity. P68 phage particles were solubilized in protein sample buffer (62.5 mM Tris-HCl [pH 6.8], 2.3% SDS, 1% mercaptoethanol, 10% glycerol, 0.01% bromophenol blue), heated for 10 min at 100°C, and then separated on SDS-phosphonoacetic acid (PAA) gels containing the autoclaved cells. After electrophoresis, the zymograms were washed for 30 min with distilled water at room temperature and then transferred into buffer containing 25 mM Tris (pH 7.5) and 0.1% Triton X-100, followed by further incubation for 16 h at 37°C. The zymograms were rinsed with distilled water, stained with 0.1% methylene blue in 0.001% KOH for 2 h at room temperature, and then destained with distilled water. Peptidoglycan hydrolase activity was detected as a clear zone on a dark blue background. Proteins of the clear zone of the zymogram were eluted using the ElutaTube extraction and dialysis kit (MBI).
Antimicrobial activity of protein 17.
The 50% inhibitory concentration of protein 17 was determined on S. aureus strain 68, which was grown in brain heart infusion (BHI) broth at 37°C. Then 5 pg/ml, 50 pg/ml, 500 pg/ml, 5 ng/ml, 50 ng/ml, or 50 μg/ml of protein 17 was added to 1 × 107 CFU. Twenty minutes after the addition of the proteins, serial dilutions were plated on BHI agar plates, and the number of CFU was determined after bacterial growth for 16 h at 37°C. The experiment was performed in triplicate. The antimicrobial spectrum of protein 17 on 35 clinical S. aureus isolates was determined similarly by growing the strains in BHI broth at 37°C until they reached an optical density at 600 nm of 0.5 (∼1 × 107 CFU). Then protein 17 (50 ng/ml) was added, and 20 min after the addition of the protein, serial dilutions were plated on BHI agar plates. The number of CFU was determined in triplicate for each strain after bacterial growth for 16 h at 37°C.
Binding of protein 17 to the bacterial surface.
A cell wall binding assay was used to determine the binding of proteins 17, 17L, and 17S, respectively, to the bacterial surface. Bacteria grown to late log phase were harvested by centrifugation and concentrated 10-fold in phosphate-buffered saline (PBS) buffer (100 mM NaCl, 3 mM KCl, 10 mM NaH2PO4, 2 mM KH2PO4, pH 7.5), and then stored on ice. One hundred microliters of cells and 100 μl (50 ng) purified protein were mixed and incubated at room temperature for 10 min. Cells were then removed from the supernatant by centrifugation (16,000 × g, 60 s). The cells were washed twice with 500 μl of PBS, and the pellet was resuspended in 50 μl PBS buffer. The same volume of protein sample buffer (62.5 mM Tris-HCl [pH 6.8], 2.3% SDS, 1% mercaptoethanol, 10% glycerol, 0.01% bromophenol blue) was added, and samples were heated for 10 min at 100°C and loaded onto an SDS-PAA gel. The presence of proteins 17, 17L, and 17S in the cell pellet (bound protein) and in the supernatant (unbound protein) was determined by a standard immunodetection procedure using antibodies raised against whole-phage particles.
To remove proteins or lipids from the cell surface, S. aureus strain 68 was subjected to various treatments. Cells from 50-ml aliquots of log-phase cultures resuspended in 250 μl PBS buffer served as starting material for the following treatments: heating (121°C, 15 min), extraction with 4% SDS or 1% Triton X-100 (50°C, 30 min), extraction with 50% phenol-50% chloroform, or digestion with 20 μg proteinase K (50°C, 30 min). After these treatments, the cells were extensively washed with PBS buffer to remove any residual detergents, solvents, or enzymes.
RESULTS
Virion protein 17 of phage P68 has muralytic activity.
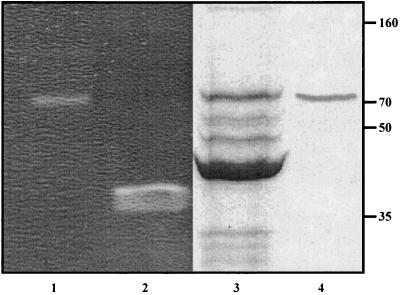
To assess whether P68 virions contain a structural component with muralytic activity, zymogram assays were performed with autoclaved S. aureus 68 cells. Upon electrophoretic separation of P68 phage proteins, the zymogram revealed a single ∼70-kDa band with muralytic activity (Fig. 1, lane 1). The corresponding phage protein was electroeluted from the clear zone and again separated on an SDS-PAA gel in parallel with purified phage P68 particles. As shown in Fig. 1, lane 4, only one protein band appeared after elution from the zymogram, which appeared to correspond in size to the minor structural protein 17 of P68 (Mr = 74,806) (45). The identity of protein 17 was confirmed by mass spectrometry of the protein eluted from the zymogram and of the corresponding phage virion protein eluted from the SDS-PAA gel. The determination of N- and C-terminal peptides by mass spectrometry further demonstrated that the protein had the predicted molecular weight, i.e., that it was not processed at either end (not shown).
FIG. 1.
Protein 17 of phage 68 has muralytic activity. 1011 phage particles (lane 1) and 10 μg of lysostaphin (lane 2) were resolved in the zymogram as described in Materials and Methods. Proteins from the clear area of the zymogram (lane 1) were eluted and resolved on a 10% SDS-PAA gel (lane 4) in parallel with purified P68 particles (lane 3). The positions of molecular mass markers (in kilodaltons) are indicated to the right.
Computer-based similarity searches revealed that protein 17 showed 27% similarity within a 493-amino-acid overlap to the minor structural protein 89 of Lactobacillus delbrueckii phage LL-H (29) and 25% similarity to the tail tip protein Tal2009 of L. lactis phage Tuc2009, belonging to the Siphoviridae. The Tal2009 has recently been shown to possess cell wall-degrading activity (16).
High MOI and addition of purified protein 17 trigger premature lysis of phage P68-infected cells.
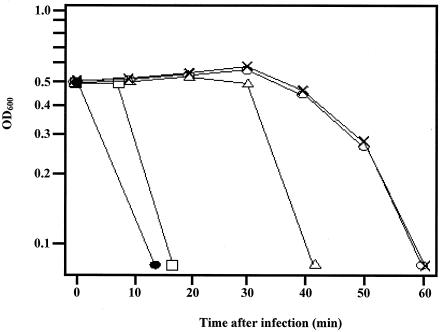
S. aureus cells infected with an MOI of 5 of phage P68 entered the lysis phase at approximately 45 to 50 min postinfection, and the culture cleared within the next 10 min (Fig. 2). In contrast, infection with a high MOI (∼100) of P68 virions caused, similarly to E. coli phage T4 (4), nearly complete lysis of S. aureus cells within 15 min of infection (Fig. 2), indicating that protein 17 may induce LO. When 50 ng/ml of protein 17 was added to P68-infected S. aureus cells 10 min prior to infection, premature lysis was observed. Premature lysis was also triggered by addition of protein 17 at 20 min postinfection, whereas the addition of protein 17 immediately before the start of vegetative lysis had no effect. These experiments suggested that P17 weakens the peptidoglycan from without.
FIG. 2.
Infection of S. aureus strain 68 with an MOI of 100 (•) and an MOI of 5 (○) at time zero culminated in lysis 15 and 40 min thereafter, respectively. Purified protein 17 (50 ng/ml) was added 10 min prior to infection (□) or 20 min postinfection (▵) with phage P68 or immediately before the onset of lysis (×). Results from a representative experiment of three independent experiments are shown. OD600, optical density at 600 nm.
Antimicrobial activity of protein 17.
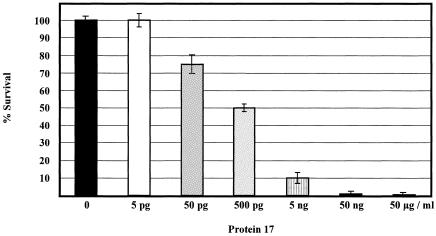
In further experiments, we tested the antimicrobial activity of purified protein 17 by exposing 1 × 107 CFU of exponentially growing S. aureus 68 cells to different concentrations of the protein for 20 min. The addition of 5 pg/ml of protein 17 to S. aureus 68 cells had no measurable effect (Fig. 3). The addition of 50 pg/ml, 500 pg/ml, 5 ng/ml, 50 ng/ml, and 50 μg/ml of protein 17 to the S. aureus culture resulted in a reduction of the plating efficiency of 25% ± 5%, 50% ± 2%, 90% ± 3%, 99% ± 1%, and 99.5% ± 0.2%, respectively (Fig. 3). From these results, we calculated the 50% inhibitory concentration for protein 17 as ∼500 pg/ml.
FIG. 3.
Antimicrobial activity of protein 17. The antimicrobial activity of protein 17 was tested by exposing 1 × 107 CFU of exponentially growing S. aureus strain 68 for 20 min to 0 pg/ml, 5 pg/ml, 50 pg/ml, 500 pg/ml, 5 ng/ml, 50 ng/ml, and 50 μg/ml of protein 17. The experiment was performed in triplicate. The error bars represent standard deviations.
The antimicrobial spectrum of protein 17 was further tested on 35 clinical isolates of S. aureus, ∼50% of which were oxacillin resistant (Table 2). Purified protein 17 (50 ng/ml) was added to exponentially growing cells (1 × 107 CFU), and serial dilutions of the cells were plated after 20 min of exposure. This treatment resulted in a reduction in the plating efficiency of ∼99% of the majority of the tested clinical S. aureus isolates (Table 2).
TABLE 2.
Host range of S. aureus phage P68 and antimicrobial spectrum of purified protein 17
| S. aureus straina | Plating of P68b | Antimicrobial activity of protein 17c | Methicillin resistanced |
|---|---|---|---|
| P1 | + | + | R |
| P2 | + | + | R |
| P11 | + | + | R |
| P16 | + | + | S |
| P17 | + | + | S |
| P21 | + | + | R |
| P30 | + | + | S |
| P34 | + | + | R |
| P35 | + | + | R |
| P4 | − | + | R |
| P6 | − | + | S |
| P8 | − | + | R |
| P9 | − | + | R |
| P10 | − | + | S |
| P13 | − | + | S |
| P14 | − | + | R |
| P15 | − | + | S |
| P18 | − | + | R |
| P19 | − | + | S |
| P23 | − | + | S |
| P24 | − | + | R |
| P25 | − | + | S |
| P26 | − | + | R |
| P29 | − | + | R |
| P31 | − | + | R |
| P32 | − | + | S |
| P5 | + | − | S |
| P27 | + | − | S |
| P3 | − | − | S |
| P7 | − | − | R |
| P12 | − | − | S |
| P20 | − | − | S |
| P22 | − | − | S |
| P28 | − | − | S |
| P33 | − | − | S |
S. aureus strains were isolated from patients.
+, phage is able to form plaques on the strain; −, no plaque formation observed.
+, protein 17 displays an antimicrobial effect. The cells (1 × 107 CFU) were first treated for 20 min with 50 ng/ml of protein 17, and then serial dilutions were plated on BHI plates. In either case, the plating efficiency was reduced by more than ∼99% compared to the nontreated control. −, no antimicrobial effect of protein 17.
Detection of methicillin resistance by oxacillin. R, the strain was considered oxacillin resistant if the interpretive zone diameter was ≤10 mm; S, when the interpretive zone diameter was ≥13 mm, the strain was considered oxacillin sensitive.
Protein 17 showed a broader antimicrobial spectrum compared to the host range of phage P68. As shown in Table 2, the phage plated only on 11 of the clinical strains, whereas protein 17 displayed activity against 26 strains. This suggested that the bacterial receptor required for phage attachment differs from the ligand required for protein 17 binding. Interestingly, the phage plated on two S. aureus isolates (P5 and P27) (Table 2) which were insensitive to protein P17. This finding could indicate that P17 is not necessarily essential for infection by P68 of every S. aureus strain.
The C terminus of protein 17 is required for binding to the cell surface.
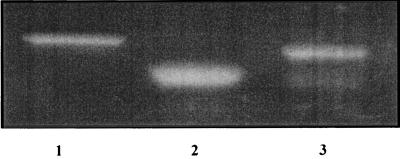
Many muralytic enzymes of gram-positive bacteria and their phage have a composite structure with the catalytic active site located in the N-terminal region, while the C-terminal part contains the target-specific binding domain (1, 12, 22, 42, 44). To test whether protein 17 has a cell surface binding domain, two C-terminally truncated variants of protein 17, lacking the C-terminal 53 aa (protein 17L) and 167 aa (protein 17S), respectively, were generated by means of recombinant DNA technology and purified. As shown in Fig. 4, both truncated versions of protein 17 displayed murein hydrolase activity in zymograms containing murein of S. aureus strain 68, demonstrating that the catalytic site of the protein 17 is located within the first 480 aa residues.
FIG. 4.
Enzymatic activity of protein 17 resides in the N-terminal part. The muralytic activities of proteins 17 (lane 1), 17S (lane 2), and 17L (lane 3), respectively, were tested in zymograms containing murein of S. aureus strain 68.
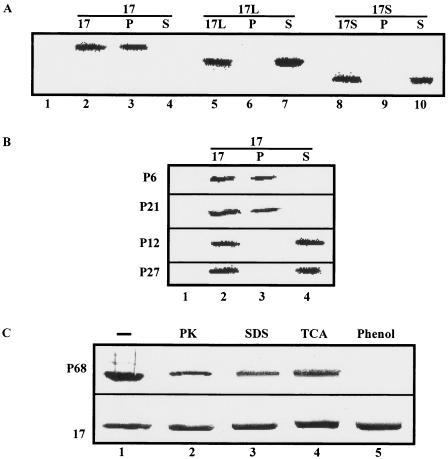
The addition of 50 ng/ml of truncated proteins 17S and 17L, respectively, to exponentially growing S. aureus 68 cells and to the clinical strains susceptible to protein 17 did not affect their viability (not shown), which led us to test whether cell targeting is abolished in 17S and 17L, respectively. Fifty nanograms of either protein 17, 17S, or 17L was added to S. aureus strain 68, and binding was assessed. As shown in Fig. 5A, both C-terminally truncated variants of protein 17 did not bind to the cells, demonstrating that the C terminus is indeed required for cell targeting of protein 17. Moreover, in agreement with the antimicrobial spectrum (Table 2), protein 17 did not bind to the clinical strains P12 and P27, whereas binding was observed to the protein 17-sensitive strains P6 and P21 (Fig. 5B).
FIG. 5.
C-terminal part of protein 17 is required for binding to the cell surface. Binding of protein 17 to the cell surface was revealed by means of immunodetection of protein 17 in Western blots using antibodies raised against whole-phage particles. Only the relevant parts of the Western blots are shown. P, the respective protein was found in the cell pellet, i.e., the protein associated with the cell surface; S, the respective protein was only found in the supernatant, i.e., did not bind to the cell surface. (A) Binding of purified proteins 17 (lanes 2 to 4), 17L (lanes 5 to 7), and 17S (lanes 8 to 10) to S. aureus 68 cells. Lane 1, protein 17 was omitted. (B) Binding of purified protein 17 to S. aureus P6, P21, P12, and P27 cells (Table 2). (C) Binding of protein 17 (lower panel) and absorption of phage P68 (upper panel) to untreated S. aureus 68 cells (lane 1) and S. aureus 68 cells treated with proteinase K (PK; lane 2), SDS (lane 3), trichloroacetic acid (TCA; lane 4), and phenol (lane 5). The cell pellet was tested for the presence of protein 17.
Ligand required for binding of protein 17 to the cell surface.
To assess the nature of the ligand required for binding of protein 17 to the bacterial surface, several treatments (heating, extraction with solvents and/or detergents, and protease digestion) were applied to remove proteins, lipids, and membrane-associated components such as lipoteichoic acids from the cell wall of S. aureus strain 68. None of these treatments had an obvious effect on binding of protein 17 (Fig. 5C). On the other hand, phenol extraction of lipoteichoic acids from S. aureus 68 cells rendered the cells resistant to phage P68 (Fig. 5C). Taken together, these experiments suggested that lipoteichoic acids serve as phage receptors and that protein 17 may target a carbohydrate ligand, which is not primarily required for phage absorption. On the other hand, they also indicated that P17 is not able to bind stably to the cell wall when it is part of the virion.
DISCUSSION
The life cycle of a bacteriophage begins with the insertion of its genetic material into the host and generally culminates in host cell lysis to allow progeny release. The peptidoglycan is a barrier for the transport of macromolecules and prevents diffusion of globular proteins larger than ∼50 kDa (5, 7). T-even phage particles have long been known to contain a lysozyme activity (protein 5) in the baseplate. It has been suggested that the lysozyme activity of protein 5 locally digests the peptidoglycan, allowing the T4 tail tube to penetrate the periplasmic space and therefore allowing the infection process to be completed (15, 34, 36). More recently, Kenny et al. (16) have demonstrated that the L. lactis phage Tuc2009 tail tip protein Tal2009 has muralytic activity. Tal2009 appears to undergo autocatalytic processing, which was attributed to its endopeptidase activity. In this study, we have shown that the virion-associated minor structural protein 17 of S. aureus phage P68 has muralytic activity. Infection of S. aureus with a high MOI of P68 (Fig. 2) resulted in lysis. In contrast, the same experiment conducted with the phage P68-resistant clinical isolates P4 and P29 did not result in lysis (not shown), although the latter were sensitive to soluble P17 (Table 2). This suggests that P68-bound P17 can only exert its activity upon adsorption of the phage to the host. Similarly, the addition of the protein 10 min prior to infection or 20 min postinfection triggered premature lysis of P68-infected S. aureus cells. Taken together, these experiments indicate that protein 17 causes a localized rupture in the cell wall, which at the initial state of infection, facilitates penetration of the phage DNA into the cytoplasm. In contrast to the Tal2009 protein, we found no evidence for a putative protease motif in P17 and did not observe any evidence for processing of the protein. Purified P17 was stable, and the MALDI-time of flight data as well as the apparent molecular weight in SDS-polyacrylamide gels agreed with that deduced from the primary protein sequence.
P68 protein P17 differs in another aspect from Tal2009 (16) in that it binds to the cell wall (Fig. 3). In gram-positive bacteria, lysozymes or lytic transglycosylases generally contain an N-terminal catalytic domain, while the C-terminal part contains the target-specific binding domain (10, 42, 44). These C-terminal domains are composed of amino acid repeats or sequence motifs required for anchoring of the enzymes to the bacterial cell surface, such as (i) an LPXTG motif for covalent linkage to the peptidoglycan cross-bridges by a sortase function (6, 41), (ii) a hydrophobic C-terminal region for anchoring to the membrane (17), (iii) sequence repeats, in particular those starting with a GW motif for binding to lipoteichoic acids (11), (iv) a bacterial S-layer homology domain for anchoring proteins to modified secondary cell wall polymers (25, 28), and (v) LysM domains, which are thought to contain a general peptidoglycan-binding motif (2). Only full-length protein 17 was shown to bind to and to exert activity against S. aureus, whereas proteins 17S and 17L displayed muralytic activity only in the zymogram assay, wherein the catalytic domain can apparently directly access the peptidoglycan substrate. Our studies indicate that protein 17 has a bipartite structure with an N-terminal catalytic site and a C-terminal surface binding domain. Nevertheless, no typical surface binding motifs were revealed for the C-terminal end. Two imperfect GW motifs were found at the C terminus of protein 17 (not shown). Since GW modules are apparently required for binding to lipoteichoic acids (11) and removal of the latter did not affect binding of protein 17 (Fig. 5C), it is, however, less likely that they are involved in targeting of protein 17 to cells. Analogous to endolysins encoded by the L. monocytogenes phage Ply118 and Ply500 (23), the ligand required for binding of protein 17 to the cell wall appears to be different from the phage receptor (Table 2 and Fig. 5C). Since the removal of proteins, lipids, and membrane-associated components from the bacterial cell wall did not affect binding of protein 17, we speculate that the cell ligand for protein 17 is a carbohydrate component, which is perhaps covalently attached to the peptidoglycan.
Staphylococcus aureus is an important human pathogen responsible for a wide variety of diseases. S. aureus is a common cause of nosocomial pneumonia and bloodstream infections as well as community-acquired infections and is known for its resistance to first-line antibiotics such as penicillin (24, 26), which underscores the need for the development of alternatives to antibiotics. Phage therapy has recently received renewed attention as an alternative for prevention and/or treatment of bacterial infections (27, 43). Alternatively, phage-encoded endolysins might provide effective agents for the control of infectious diseases caused by gram-positive bacteria (9), the peptidoglycan of which is externally accessible. Like phage therapy, the specificity of phage-encoded endolysins offers the advantage of targeted killing of pathogens while leaving the beneficial microflora intact. For instance, the muramidase Cpl-1 derived from a pneumococcal phage was shown to be efficient in clearing pneumococci from the nasopharyngeal mucosa when applied topically but also killed S. pneumoniae in the bloodstream and significantly extended the survival of the infected mice (21). Protein 17 displayed activity against clinical S. aureus isolates, some of which were methicillin resistant. As apparent from Table 2, these agents may have an even broader activity spectrum than the phage from which they are derived. It seems, therefore, conceivable to develop virion-associated murein hydrolases toward effective antimicrobial agents to combat gram-positive pathogens in a similar way as shown for phage-derived endolysins by Fischetti (9).
Acknowledgments
We are grateful to M. Loessner, Technical University of Zürich, for critical reading of the manuscript.
REFERENCES
- 1.Baba, T., and O. Schneewind. 1996. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 15:4789-4797. [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 3.Bernhardt, T. G., I. N. Wang, D. K. Struck, and R. Young. 2001. A protein antibiotic in the phage Qβ virion: diversity in lysis targets. Science 292:2326-2329. [DOI] [PubMed] [Google Scholar]
- 4.Delbrück, M. 1940. The growth of bacteriophage and lysis of the host. J. Gen. Physiol. 23:643-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demchick, P., and A. L. Koch. 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178:768-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhar, G., K. F. Faull, and O. Schneewind. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 39:3725-3733. [DOI] [PubMed] [Google Scholar]
- 7.Dijkstra, A. J., and W. Keck. 1996. Peptidoglycan as a barrier to transenvelope transport. J. Bacteriol. 178:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emrich, J., and G. Streisinger. 1968. The role of phage lysozyme in the life cycle of phage T4. Virology 36:387-391. [DOI] [PubMed] [Google Scholar]
- 9.Fischetti, V. A. 2003. Novel method to control pathogenic bacteria on human mucous membranes. Ann. N. Y. Acad. Sci. 987:207-214. [DOI] [PubMed] [Google Scholar]
- 10.Hermoso, J. A., B. Monterroso, A. Albert, B. Galán, O. Ahrazem, P. García, M. Martínez-Ripoll, J. L. García, and M. Menéndez. 2003. Structural basis for selective recognition of pneumococcal cell wall by modular endolysin from phage Cp-1. Structure 11:1239-1249. [DOI] [PubMed] [Google Scholar]
- 11.Jonquières, R., H. Bierne, F. Fiedler, P. Gounon, and P. Cossart. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of Gram-positive bacteria. Mol. Microbiol. 34:902-914. [DOI] [PubMed] [Google Scholar]
- 12.Joris, B., S. Englebert, C. P. Chu, R. Kariyama, L. Daneo-Moore, G. D. Shockman, and J. M. Ghuysen. 1992. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 70:257-264. [DOI] [PubMed] [Google Scholar]
- 13.Kanamaru, S., N. C. Gassner, Y. Nanzhang, S. Takeda, and F. Arisaka. 1999. The C-terminal fragment of the precursor tail lysozyme of bacteriophage T4 stays as a structural component of the baseplate after cleavage. J. Bacteriol. 181:2739-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao, S. H., and W. H. McClain. 1980. Roles of bacteriophage T4 gene 5 and gene s products in cell lysis. J. Virol. 34:104-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao, S. H., and W. H. McClain. 1980. Baseplate protein of bacteriophage T4 with both structural and lytic functions. J. Virol. 34:95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenny, J. G., S. McGrath, G. F. Fitzgerald, and D. van Sinderen. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J. Bacteriol. 186:3480-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kocks, C., E. Gouin, M. Tabouret, P. Berche, H. Ohayon, and P. Cossart. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521. [DOI] [PubMed] [Google Scholar]
- 18.Koonin, E. V., and K. E. Rudd. 1994. A conserved domain in putative bacterial and bacteriophage transglycosylases. Trends Biochem. Sci. 19:106-107. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Lepeuple, A. S., E. Van Gemert, and M. P. Chapot-Chartier. 1998. Analysis of the bacteriolytic enzymes of the autolytic Lactococcus lactis subsp. cremoris strain AM2 by renaturing polyacrylamide gel electrophoresis: identification of a prophage-encoded enzyme. Appl. Environ. Microbiol. 64:4142-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loeffler, J. M., S. Djurkovic, and V. A. Fischetti. 2003. Phage lytic enzyme Cpl-1 as a novel antimicrobial for pneumococcal bacteremia. Infect. Immun. 71:6199-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loessner, M. J., S. Gaeng, G. Wendlinger, S. K. Maier, and S. Scherer. 1998. The two-component lysis system of Staphylococcus aureus bacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol. Lett. 162:265-274. [DOI] [PubMed] [Google Scholar]
- 23.Loessner, M. J., K. Kramer, F. Ebel, and S. Scherer. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335-349. [DOI] [PubMed] [Google Scholar]
- 24.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 25.Lupas, A., H. Engelhardt, J. Peters, U. Santarius, S. Volker, and W. Baumeister. 1994. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J. Bacteriol. 176:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merril, C. R., D. Scholl, and S. L. Adhya. 2003. The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug Discov. 2:489-497. [DOI] [PubMed] [Google Scholar]
- 28.Mesnage, S., E. Tosi-Couture, and A. Fouet. 1999. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol. Microbiol. 31:927-936. [DOI] [PubMed] [Google Scholar]
- 29.Mikkonen, M., and T. Alatossava. 1994. Characterization of the genome region encoding structural proteins of Lactobacillus delbrueckii subsp. lactis bacteriophage LL-H. Gene 151:53-59. [DOI] [PubMed] [Google Scholar]
- 30.Mindich, L., and J. Lehman. 1979. Cell wall lysin as a component of the bacteriophage φ6 virion. J. Virol. 30:489-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moak, M., and I. J. Molineux. 2000. The role of the lytic transglycosylase motif of bacteriophage T7 in the initiation of infection. Mol. Microbiol. 37:345-355. [DOI] [PubMed] [Google Scholar]
- 32.Moak, M., and I. J. Molineux. 2004. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51:1169-1183. [DOI] [PubMed] [Google Scholar]
- 33.Molineux, I. J. 2001. No syringes please, ejection of T7 DNA from the virion is enzyme-driven. Mol. Microbiol. 40:1-8. [DOI] [PubMed] [Google Scholar]
- 34.Mosig, G., W. Lin, J. Franklin, and W. H. Fan. 1989. Functional relationships and structural determinants of two bacteriophage T4 lysozymes: a soluble (gene e) and a baseplate-associated (gene 5) protein. New Biol. 1:171-179. [PubMed] [Google Scholar]
- 35.Mukai, F., G. Streisinger, and B. Miller. 1967. The mechanism of lysis in phage T4-infected cells. Virology 33:398-404. [DOI] [PubMed] [Google Scholar]
- 36.Nakagawa, H., F. Arisaka, and S. Ishii. 1985. Isolation and characterization of the bacteriophage T4 tail-associated lysozyme. J. Virol. 54:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Committee for Clinical Laboratory Standards. 1993. Approved standards M2-A5. Performance standards for antimicrobial disk susceptibility tests. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 38.Rydman, P. S., and D. H. Bamford. 2000. Bacteriophage PRD1 DNA entry uses a viral membrane-associated transglycosylase activity. Mol. Microbiol. 37:356-363. [DOI] [PubMed] [Google Scholar]
- 39.Rydman, P. S., and D. H. Bamford. 2002. The lytic enzyme of bacteriophage PRD1 is associated with the viral membrane. J. Bacteriol. 184:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 42.Sheehan, M. M., J. L. Garcia, R. López, and P. Garcia. 1997. The lytic enzyme of the pneumococcal phage Dp-1: a chimeric lysin of intergeneric origin. Mol. Microbiol. 25:717-725. [DOI] [PubMed] [Google Scholar]
- 43.Sulakvelidze, A., Z. Alavidze, and J. G. Morris, Jr. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Usobiaga, P., F. J. Medrano, M. Gasset, J. L. Garcia, J. L. Saiz, G. Rivas, J. Laynez, and M. Menendez. 1996. Structural organization of the major autolysin from Streptococcus pneumoniae. J. Biol. Chem. 271:6832-6838. [DOI] [PubMed] [Google Scholar]
- 45.Vybiral, D., M. Takáč, M. Loessner, A. Witte, U. Ahsen, and U. Bläsi. 2003. Complete nucleotide sequence and molecular characterization of two lytic Staphylococcus aureus phages: 44AHJD and P68. FEMS Microbiol. Lett. 219:275-283. [DOI] [PubMed] [Google Scholar]
- 46.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young, R., and U. Bläsi. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol. Rev. 17:191-205. [DOI] [PubMed] [Google Scholar]
- 48.Young, R., I. N. Wang, and W. D. Roof. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8:120-128. [DOI] [PubMed] [Google Scholar]