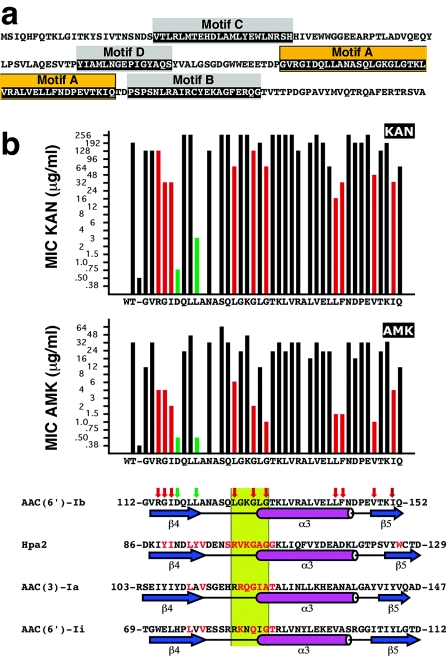
FIG. 1.
(a) Amino acid sequence of the AAC(6′)-Ib protein. The boxes indicate the conserved motifs A through D. Orange-highlighted amino acids indicate motif A. (b) Susceptibilities of E. coli harboring plasmids containing the mutant derivatives of AAC(6′)-Ib and alignment of the motif A with the amino acid sequences of other members of the GNAT superfamily. The MICs of KAN and AMK are plotted on top, the vertical axis shows the antibiotic concentrations as they are in the E-strip, and the bars show the results for the controls (plasmidless E. coli and E. coli harboring the wild-type pJHCMW1) and each substitution with alanine. The bottom part of panel b shows the alignment of the amino acid sequences of the motif A of each of the indicated GNAT proteins. The amino acid sequence of motif A in Hpa2 (accession number AY558056), AAC(3)-Ia (accession number S68049) (9), and AAC(6′)-Ii (accession number L12710) (4), as well as the structures of the regions as determined before (1, 22, 23), are shown aligned and compared to the amino acid sequence and the predicted secondary structure of the motif A of AAC(6′)-Ib (accession number AF479774) (19). Amino acids in red are involved in contacts with the cofactor acetyl-CoA (1, 3, 22). The β strands and α helix are indicated below each sequence; they are named as described by Dyda et al. (7). The green box identifies the conserved region Q/RxxGxG/A. The red vertical arrows on top of the AAC(6′)-Ib amino acid sequence indicate substitutions that reduced the MIC of AMK to 8 μg/ml or less while the MICs of KAN were not significantly reduced or reduced to no less than 16 μg/ml. The green vertical arrows indicate substitutions that resulted in MICs of AMK comparable to that shown by the plasmidless control and in MICs of KAN that were ≤3 μg/ml.