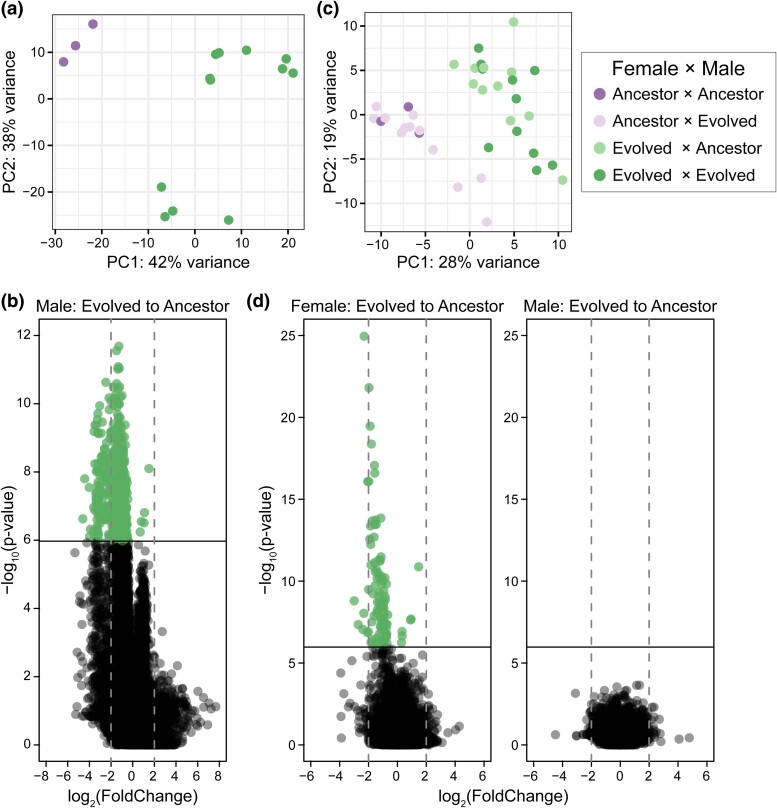
Fig. 1.
Differential gene expression is partitioned by ancestral (purple) and evolved (green) individuals. a) PCA of the variance stabilized data set partitions male generation along PC1. b) Differential gene expression was analyzed based on male generation and experimental evolution replicate. Only the male term contributed to gene expression changes (supplementary file S1, Supplementary Material online). Genes with a significant change in expression from ancestral to evolved males (shown in green) were determined using a Bonferroni cutoff. Positive expression changes correspond to upregulation in evolved males, while negative expression changes correspond to downregulation in evolved males. c) PCA of the variance stabilized data set partitions female generation along PC1. d) Differential gene expression was analyzed based on female generation, male generation, and experimental evolution replicate. Only the female term contributed to gene expression changes. Genes with a significant change in expression from ancestral to evolved females (shown in green) were determined using a Bonferroni cutoff. Positive expression changes correspond to upregulation in evolved females, while negative expression changes correspond to downregulation in evolved females.