Abstract
We found ampicillin- and imipenem-resistant isolates of vanA-possessing Enterococcus faecalis with MICs of 8 to 16 μg/ml and 4 to 32 μg/ml, respectively. There have been few reports about penicillin- and imipenem-resistant E. faecalis. Two mechanisms of beta-lactam resistance in E. faecalis, the production of beta-lactamase and the overproduction of penicillin-binding proteins (PBPs), have been reported. The resistant isolates in the current study did not produce any beta-lactamases and analysis of the PBPs showed no overproduction. However, the affinities of PBP4 for beta-lactams in the resistant strains were lower than those of susceptible strains but the affinities of other PBPs for beta-lactams did not change. Accordingly, whole pbp4 fragments from these resistant isolates were sequenced. Two amino acid substitutions at positions 520 and 605 were observed in the highly resistant strains compared to the susceptible ones, Pro520Ser and Tyr605His, and a single Tyr605His amino acid substitution was found in the low-resistance strains. These two point mutations exist in the region between the active-site-defining motifs SDN and KTG of the penicillin-binding domain, the main target of beta-lactams. A strong correlation was seen between these substitutions and decreasing affinities of PBP4 to beta-lactams. In E. faecalis, resistance due to mutations in PBPs has not been reported, though it has in Enterococcus faecium. Our results suggest that development of high-level resistance to penicillins and imipenem depends on point mutations of PBP4 at positions 520 and 605.
It has been reported that the frequency of nosocomial enterococcal infections has increased markedly in the past two decades in the United States (25). Enterococci account for over 10% of nosocomial bloodstream infections and have been associated with an attributable mortality of over 30% in the United States (5). The impact of multidrug-resistant pathogens has been augmented by the emergence of vancomycin-resistant enterococci (VRE). VRE were first isolated in Europe in 1986 (16, 36) and then in the United States in 1987 (33). The prevalence of nosocomial VRE in the United States increased from 0.3% in 1989 to 7.9% in 1993 (3). In the United States, VRE currently account for 20 to 25% of all enterococcal isolates in intensive care units, according to the National Nosocomial Infections Surveillance System, 1989 to 1998 (25). In contrast to the findings in the United States, clinical isolates of VRE in European hospitals have been rather infrequent (39). In Japan, a few cases have been reported since the first reported case of VRE infection in 1997 (13). Nosocomial VRE infections of 36 inpatients in Kitakyusyu City in Japan were reported in 2002. Most VRE are Enterococcus faecium in the United States but Enterococcus faecalis is isolated more than E. faecium among VRE in Japan (22).
Enterococci are intrinsically resistant to cephems (2, 9, 18) and most enterococcal strains such as those of E. faecium and E. avium, with the exception of E. faecalis, are also resistant to penicillins and imipenem. However, there are few reports about penicillin- and imipenem-resistant E. faecalis. Two mechanisms for beta-lactam resistance in E. faecalis have been reported. One is production of beta-lactamase to hydrolyze beta-lactams (24) and the other is the overproduction of penicillin-binding proteins (PBPs) that bind the agents (10).
We found ampicillin- and imipenem-resistant isolates of E. faecalis carrying vanA among clinical specimens. We studied the mechanisms of resistance to penicillin and imipenem in these isolates.
MATERIALS AND METHODS
Bacterial strains.
The E. faecalis strains used in this study are listed in Table 1. These seven strains were isolated from different inpatients at three hospitals between 1998 and 2000 in Kitakyusyu, Japan. All isolates were identified as E. faecalis using the Vitek system (bioMerieux, France). The vancomycin resistance genes, vanA and vanB, were detected by PCR using specific primers (7, 15). SEF 96, ATCC 29212, SVR 250, and SVR 251 were ampicillin- and imipenem-susceptible strains. SVR 34 and SVR 138 were ampicillin- and imipenem-insensitive strains. SVR 1110 and SVR 1119 were ampicillin- and imipenem-resistant strains.
TABLE 1.
Bacterial strains and MIC values of selected agents
| Strain | Date of isolation (day.mo.yr) | Specimen and relevant characteristics | MIC (μg/ml)a
|
Beta-lactamase produced | |||
|---|---|---|---|---|---|---|---|
| AMP | IPM | CTX | VCM | ||||
| Enterococcus faecalis | |||||||
| SEF96 | 21.04.2000 | Clinical isolate from urine | 1 | 1 | >256 | 1 | − |
| ATCC 29212 | Type strain | 1 | 1 | 256 | 4 | − | |
| SVR 34 | 15.02.2000 | Clinical isolate with vanB from urine | 8 | 4 | >256 | 32 | − |
| SVR 138 | 27.05.2002 | Clinical isolate with vanA from stool | 8 | 8 | >256 | >512 | − |
| SVR 250 | 19.07.2002 | Clinical isolate with vanA from urine | 1 | 0.5 | 64 | >512 | − |
| SVR 251 | 19.07.2002 | Clinical isolate with vanA from urine | 1 | 0.5 | 64 | >512 | − |
| SVR 1110 | 02.09.2000 | Clinical isolate with vanA from urine | 16 | 32 | >256 | >512 | − |
| SVR 1119 | 01.12.1998 | Clinical isolate with vanA from urine | 16 | 32 | >256 | >512 | − |
| SVR 1119S | ΔvanA; spontaneous mutant derived from SVR 1119 | 16 | 32 | >256 | 1 | ||
| Escherichia coli | |||||||
| ATCC 35218 | Beta-lactamase-positive strain | + | |||||
Abbreviations: AMP, ampicillin; IPM, imipenem; CTX, cefotaxime; VCM, vancomycin.
SVR 1119S, without vanA, was selected from SVR 1119 using a serial passage method in vancomycin-free BHI broth (Eiken Chemical Co., Ltd., Japan) and then a replica planting method on BHI agar (17, 31).
Antimicrobial agents and MIC determination.
Antimicrobial agents were obtained as follows: ampicillin and vancomycin, Nacalai Tesque, Inc.; imipenem, Banyu; and cefotaxime, Sigma Chemical Co. MICs were determined on Mueller-Hinton agar by a serial dilution method (29).
Beta-lactamase activity.
For hydrolysis studies, E. faecalis isolates were grown overnight in BHI broth, harvested, washed once in 50 mM sodium phosphate buffer (pH 7.0), and resuspended in 0.1 volume of sodium phosphate buffer. The suspension was sonically disrupted using an Ultrasonic disruptor UD-201 (TOMY, Tokyo, Japan) and debris was removed by centrifugation. The supernatant was tested for beta-lactamase activity with nitrocefin (Oxoid, Ltd., United Kingdom). Escherichia coli ATCC 35218 was used as a positive control for beta-lactamase activity (27).
Isolation of cytoplasmic membranes.
Bacterial cytoplasmic membranes were prepared following the method of Spratt et al. (35). Briefly, cells were grown in BHI broth at 37°C overnight, collected in late exponential growth phase, and disrupted by sonication on ice. Residual cells were removed by centrifugation; protein concentration in the remaining supernatant was estimated using Lowry's method (20). Membranes were then stored at −80°C in 50 mM sodium phosphate, pH 7.0, containing 10 mM MgCl2.
Analysis of PBPs.
Bocillin FL (Molecular Probes, Inc., Eugene, Oregon), a commercially available fluorescent penicillin, was used to detect PBPs in bacterial membrane preparations following the method of Zhao et al. (40) with some modifications. Briefly, 20 μl of a 20 mg/ml membrane protein suspension was incubated with 15 μl of 0.05 mM Bocillin FL for 10 min at 30°C. The proteins were then solubilized with sarcosyl, 180 mg/ml penicillin G was added, and the membrane fraction was obtained by centrifugation. Labeled membrane proteins were resolved by gel electrophoresis and visualized with UV light (ATTO AB1500, wavelength 312 nm).
PBP competition assay.
A competition assay was also used to analyze the PBPs from these isolates. Membrane proteins (20 mg/ml) were incubated with increasing concentrations of ampicillin or imipenem (from 0.0625 to 32 μl/ml) for 10 min at 30°C. Samples were then incubated with 0.05 mM Bocillin FL for another 10 min at 30°C and analyzed as described above. The 50% inhibitory concentration (IC50) was taken to be the minimum concentration of unlabeled antibiotic which reduced binding of the Bocillin FL to a PBP by more than 50%.
Amplification and sequence analysis of the pbp4 gene.
The whole pbp4 gene of E. faecalis used was amplified by PCR using specific primers (Table 2). The PCR products were subjected to direct sequencing reactions by the dideoxy chain termination method with an ABI PRISM 377 DNA sequencer (PE Biosystems).
TABLE 2.
Primers designed for sequencing of PBP4 genes
| Primer | Sequence | Reference |
|---|---|---|
| EF PBP4 140′F | 5′CAACGAAAGCCTGATGAAATGG3′ | This study |
| EF PBP4 1043F | 5′CGATTGACAGTGTACAACAACAAGC3′ | This study |
| EF PBP4 1132R | 5′AATCGCCTTTTTGAGGATCGG3′ | This study |
| EF PBP4 2130R | 5′CGCTTCATTGTAGCACACTTTCCTTTTTC3′ | 4 |
Nucleotide sequence accession numbers.
The NCBI accession numbers for the nucleotide sequences of the pbp4 genes of E. faecalis ATCC 29212, SVR 251, SVR 1110, and SVR 1119 are AY571368, AY571365, AY571366, and AY571367, respectively.
RESULTS
Susceptibility to beta-lactam antibiotics.
Table 1 shows the susceptibility of the strains used in this study to ampicillin, imipenem, cefotaxime, and vancomycin. The MICs of ampicillin and imipenem against E. faecalis SEF96, ATCC 29212, SVR 250, and SVR 251, which were ampicillin-susceptible strains, were 1 μg/ml and 0.5 to 1 μg/ml, respectively. The MICs of ampicillin and imipenem against E. faecalis SVR 34 and SVR 138, which were ampicillin-insensitive strains, were 8 and 4 to 8 μg/ml, respectively. The MICs of ampicillin and imipenem against E. faecalis SVR 1110 and SVR 1119, which were ampicillin-resistant strains, were 16 and 32 μg/ml, respectively.
The susceptibility of SVR 1119S, which was selected as a vanA deletion mutant from SVR 1119, was the same as the susceptibility of SVR 1119 to antibiotics except vancomycin.
Beta-lactamase activity.
None of the strains used in this study hydrolyzed nitrocefin with the exception of a positive control strain, Escherichia coli ATCC 35218.
PBP expression.
We used a fluorescent penicillin (Bocillin FL) to detect PBP in membrane fractions prepared from all of the isolates. The PBPs of all strains were expressed as the same five bands that corresponded to proteins of approximately 107 kDa, 81 kDa, 76 kDa, 72 kDa, and 67 kDa. The amount of the corresponding PBP of each strain was almost the same. So, overproduction of any PBPs by any strain was not seen.
PBP competition assay.
We performed competition assays to measure the affinity of each PBP for ampicillin or imipenem. The IC50 (μg/ml) is expressed as the minimum concentration of antibiotic required to reduce Bocillin FL binding to the PBP by more than 50%. There were obvious differences in PBP4 saturation for ampicillin or imipenem between the susceptible strain, SVR 250, and the resistant strain, SVR 1110. The IC50 for PBP4 of the susceptible strains for both antibiotics was obviously lower than that of the resistant strains (Table 3).
TABLE 3.
Inhibition of binding of Bocillin FL to PBPs by two beta-lactams
| Drug | E. faecalis strain | IC50a (μg/ml)
|
MIC (μg/ml) | |||
|---|---|---|---|---|---|---|
| PBP1 | PBP3 | PBP4 | PBP5 | |||
| Ampicillin | SEF96 | 1 | 0.5 | 0.25 | >16 | 1 |
| ATCC 29212 | 1 | 0.5 | 0.25 | >16 | 1 | |
| SVR 250 | 2 | 0.5 | 1 | >8 | 1 | |
| SVR 251 | 2 | 0.5 | 0.25 | >8 | 1 | |
| SVR 34 | 2 | 0.5 | 4 | >32 | 8 | |
| SVR 138 | 2 | 0.5 | 4 | >32 | 8 | |
| SVR 1110 | 1 | 0.5 | 8 | >32 | 16 | |
| SVR 1119 | 1 | 0.25 | 8 | >32 | 16 | |
| Imipenem | SEF96 | 0.5 | 0.25 | 0.25 | >16 | 1 |
| ATCC 29212 | 0.5 | 0.25 | 0.125 | >16 | 1 | |
| SVR 250 | 1 | 0.5 | 0.25 | >8 | 0.5 | |
| SVR 251 | 0.5 | 0.25 | 0.5 | >8 | 0.5 | |
| SVR 34 | 0.5 | 0.25 | 4 | >32 | 4 | |
| SVR 138 | 0.5 | 0.25 | 4 | >32 | 4 | |
| SVR 1110 | 1 | 0.5 | 32 | >32 | 32 | |
| SVR 1119 | 1 | 0.25 | 16 | >32 | 32 | |
IC50, concentration of unlabeled antibiotic which reduces binding of Bocillin FL to a PBP by more than 50%.
The IC50 of ampicillin against PBP4 of the resistant strains (E. faecalis SVR 1110 and SVR 1119) was 8 μg/ml, 8 to 32 times higher than those against susceptible strains (SEF96, ATCC 29212, SVR 250, and SVR 251). The IC50s of ampicillin against the insensitive strains were 4 to 16 times higher than those against susceptible strains.
The IC50s of imipenem against PBP4 of the resistant strains (E. faecalis SVR 1110 and SVR 1119) were 16 and 32 μg/ml, respectively, 32 to 256 times higher, respectively, than those against susceptible strains (SEF96, ATCC 29212, SVR 250, and SVR 251). Those of imipenem against insensitive strains were 8 to 32 times higher than those against susceptible strains.
The IC50s of ampicillin and imipenem against PBP4 of all strains had a positive correlation with the respective MICs. There were no differences between the IC50s of ampicillin and imipenem against any of the other PBPs of susceptible or resistant strains.
Sequencing of PBP4.
We amplified and sequenced the entire pbp4 gene, corresponding to amino acids 1 to 680, to identify mutations that may account for the differences in the affinity of this protein between the susceptible and resistant strains for beta-lactams. The beta-lactam-resistant strains (SVR 1110 and SVR 1119) had two amino acid substitutions encoded by the pbp4 gene compared to the reference strain (SEF96), Pro520Ser and Tyr605His (Table 4). The insensitive strains (SVR 34 and SVR 138) had a single Tyr605His amino acid substitution encoded by pbp4. All substitutions were due to point mutations. Point mutations were also seen in the susceptible strains ATCC 29212 (Val359Ala) and SVR 250 and SVR 251 (Tyr50Ile) compared with SEF96, but these point mutations had no effect on the MIC of beta-lactams for these strains.
TABLE 4.
Amino acid alterations in strains in this study compared with E. faecalis SEF96
| Strain | MICs (μg/ml), ampicillin/imipenem | IC50 (μg/ml), ampicillin/imipenem | Alteration at position:
|
|||
|---|---|---|---|---|---|---|
| 50 | 369 | 520 | 605 | |||
| SEF96 | 1/1 | 0.25/0.25 | Tyr | Val | Pro | Tyr |
| ATCC 29212 | 1/1 | 0.25/0.125 | Ala | |||
| SVR 250 | 1/0.5 | 1/0.25 | Ile | |||
| SVR 251 | 1/0.5 | 0.25/0.5 | Ile | |||
| SVR 34 | 8/4 | 4/4 | His | |||
| SVR 138 | 8/4 | 4/4 | His | |||
| SVR 1110 | 16/32 | 8/32 | Ser | His | ||
| SVR 1119 | 16/32 | 8/32 | Ser | His | ||
DISCUSSION
We found ampicillin- and imipenem-resistant isolates of E. faecalis carrying vanA. There have been few reports about ampicillin- and imipenem-resistant E. faecalis. MICs for ampicillin and imipenem against both SVR 1119 and SVR 1119S, which was cured of a vanA-carrying plasmid from SVR 1119, were identical. Therefore, vanA was not involved in the development of penicillin resistance in these strains.
Two mechanisms of beta-lactam resistance have been reported in enterococci. Beta-lactamase production is one of these resistance mechanisms. Murray et al. have reported beta-lactamase-mediated penicillin resistance in E. faecalis (26-28). However, with the exception of a few reports (14, 23), enterococci do not produce beta-lactamases. In fact, the penicillin- and imipenem-resistant E. faecalis isolates in this study did not produce any beta-lactamases.
The other mechanism of resistance is a change in the affinity of penicillin-binding proteins for beta-lactams or overproduction of specific penicillin-binding proteins. When penicillin binds to a specific PBP at a certain fixed ratio, cell growth stops or cells die; these PBPs are called low-affinity PBPs. Alteration of the affinity of a PBP could have an important role in the mechanism of resistance to beta-lactams. Williamson et al. showed that at least five PBPs have been found in E. faecalis by labeling membranes with radioactive penicillin (38), and five PBP genes were found in the genome of E. faecalis V583 (GenBank NC004668). We also identified five PBPs in the E. faecalis strains used in this study when membranes were labeled with Bocillin FL. The roles of PBPs in penicillin resistance are partially known in some Enterococcus species, such as E. faecium, E. hirae, and E. faecalis.
Fontana et al. have reported that PBP 1 and -2 were not detected when E. faecalis was cultured at 32°C. These two PBPs were not essential for survival or cell growth of E. faecalis (11). Mutants of E. faecium lacking PBP5 were hypersensitive to penicillin (8). Overproduction of low-affinity PBPs has also been associated with development of penicillin resistance in enterococcal species (E. faecium, E. faecalis, E. avium, etc.) by studies of in vitro-selected penicillin-resistant mutants or penicillin-resistant clinical isolates (1, 12, 37, 41).
In E. faecium, ampicillin resistance has been shown to arise from not only overproduction but also amino acid substitutions of PBP5 (19, 42). Ligozzi et al. showed that in E. faecium, four amino acid changes in PBP5 occurred between an ampicillin-sensitive strain, E. faecium 28S (0.5 μg/ml), and an ampicillin-resistant strain, E. faecium 9439 (128 μg/ml) (19). Zorzi et al. showed that development of intermediate resistance (benzylpenicillin MICs of 6 to 64 μg/ml) in some E. faecium strains could be explained by the overproduction of unchanged PBP5 and high-level resistance of two E. faecium strains, EFM-1 and H80721 (MICs, 90 and 512 μg/ml) was related to different amino acid substitutions yielding low-affinity PBP5s that were synthesized in small quantities (42).
On the other hand, the synthesis of PBP5 in some strains of E. hirae and E. faecium was reported to be under the control of a repressor-encoding gene, psr (for PBP5 synthesis repressor), which is localized immediately upstream of the pbp5 gene (18, 21, 32). E. faecalis JH2-2r (benzylpenicillin MIC, 75 μg/ml) was selected from E. faecalis JH2-2 by successive growth on BHI plates containing increasing concentrations of benzylpenicillin. This strain showed overproduction of PBP4, which is regarded as a low-affinity PBP. But, in their study, the cloning and sequencing of that psr-like gene from both E. faecalis JH2-2 and JH2-2r indicated that they were identical (4). No overproduction of PBPs, including PBP4, of the four imipenem-resistant strains in this study was observed. The beta-lactam affinities of PBP4 of these imipenem-resistant strains were lower than those of imipenem-susceptible strains, but the affinities of other PBPs did not change. The affinities of PBP4 of E. faecalis to ampicillin and imipenem were proportionally related to the MICs.
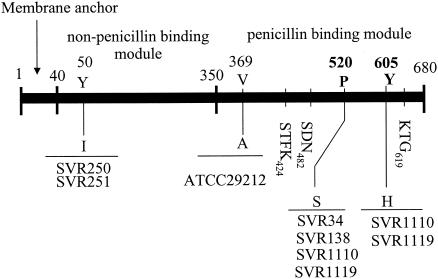
We showed that imipenem resistance in E. faecalis depends on low-affinity PBP4. Therefore, we focused on amino acid alterations of PBP4 regardless of a psr (or psr-like) gene in enterococci. Signoretto et al. have described the domain structure of the E. faecalis PBP5 as a membrane protein composed of 679 amino acids with three distinct modules. One is a penicillin-binding module (amino acids 350 to 679) located towards the C terminus, in which the three typical penicillin-binding motifs, SXXK, SDN, and KTG, were identified. The others are a non-penicillin-binding domain (amino acids 40 to 349) and an uncleaved N-terminal segment that acts as a membrane-spanning domain (amino acids 1 to 39) (34) (Fig. 1). Similar PBP arrangements could be seen in E. faecium and E. hirae, although the number of amino acids differs slightly (6, 19, 41). In E. faecium strains, it has been shown in earlier studies (10, 18, 19) that resistance to ampicillin or benzylpenicillin was associated with amino acid substitutions in the region between the active-site-defining motifs SDN and KTG of the penicillin-binding domain.
FIG. 1.
Amino acid substitutions in pbp4 of E. faecalis strains. The homology boxes (STFK with active-site serine SDN and KTG) are indicated above. The positions at which resistant and low-resistant strains are altered from the sequence of the reference strain E. faecalis SEF96 are indicated.
In our study, 605-Thr was changed to His in the insensitive strains (SVR 34 and SVR 138). Simultaneously, 520-Pro was replaced with Ser, and the same alteration at position 605 as in the insensitive strains occurred in the resistant strains (SVR 1110 and SVR 1119). The simultaneous occurrence of these two point mutations at positions 520 and 605 might be the cause of the high-level resistance to beta-lactam antibiotics. E. faecalis SEF96, which is a penicillin-susceptible strain, and JH2-2 have completely identical PBP4 protein sequences. Therefore, E. faecalis SEF96 was used as a reference strain.
Substitution of Ala for Val at position 369 in E. faecalis ATCC 29212 did not change susceptibility to ampicillin and imipenem. A point mutation between imipenem-susceptible VRE strains (SVR 250 and SVR 251) and E. faecalis ATCC 29212 produced almost no resistance to beta-lactams, either. Because all of these amino acid alterations occurred in the non-penicillin-binding module or outside the region between the active-site-defining motifs SDN and KTG, they were not likely to change the affinity of PBP4 and susceptibility of those strains.
We conclude that development of high-level resistance to beta-lactams such as imipenem and ampicillin depends on amino acid alterations of PBP4 at both positions 520 and 605. This is the first report regarding amino acid alterations of PBP4 in E. faecalis which are able to lower the affinities for and susceptibilities to beta-lactams.
REFERENCES
- 1.Al-Obeid, S., L. Gutmann, and R. Willamson. 1990. Modification of penicillin-binding proteins of penicillin resistant mutants of different species of enterococci. J. Antimicrob. Chemother. 26:613-618. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson, B. 1986. Species incidence and trends of susceptibility to antibiotics in the United States and other countries: MIC and MBC, p. 162-165. In V. Lorian (ed.), Antibiotics in laboratory medicine, 2nd ed. Williams and Wilkins Baltimore, Md.
- 3.Centers for Disease Control and Prevention. 1993. Nosocomial enterococci resistant to vancomycin-United States, 1989-1993. JAMA 270:1796. [PubMed] [Google Scholar]
- 4.Duez, C., W. Zorzi, F. Sapunaric, A. Amoroso, I. Thamm, and J. Coyette. 2001. The penicillin resistance of Enterococcus faecalis JH2-2r results from an overproduction of the low-affinity penicillin-binding protein PBP4 and does not involve a psr-like gene. Microbiology 147:2561-2569. [DOI] [PubMed] [Google Scholar]
- 5.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 6.El Kharrohbi, A., P. Jacques, G. Piras, J. Van Beeumen, J. Coyette, and J. M. Ghuysen. 1991. The Enterococcus hirae R40 penicillin binding protein 5 and the methicillin-resistant Staphylococcus aureus penicillin binding protein 2′ are similar. Biochem. J. 280:463-469. [PMC free article] [PubMed] [Google Scholar]
- 7.Elsayed, S., N. Hamilton, D. Boyd, and M. Mulvey. 2001. Improved primer design for multiplex PCR analysis of vancomycin-resistant Enterococcus spp. J. Clin. Microbiol. 39:2367-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontana. R., A. Grossato, L. Lossi, Y. R. Cheng, and G. Satta. 1985. Transition from resistance to hypersusceptibility to beta-lactam antibiotics associated with loss of a low-affinity penicillin binding protein in a Streptococcus faecium mutant highly resistant to penicillin. Antimicrob. Agents Chemother. 28:678-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontana, R., G. Amalfitano, L. Rossi, and G. Satta. 1992. Mechanisms of resistance to growth inhibition and killing by beta-lactam antibiotics in enterococci. Clin. Infect. Dis. 15:486-489. [DOI] [PubMed] [Google Scholar]
- 10.Fontana, R., M. Ligozzi, F. Pittaluga, and G. Satta. 1996. Intrinsic penicillin resistance in enterococci. Microb. Drug Resist. 2:209-213. [DOI] [PubMed] [Google Scholar]
- 11.Fontana, R., P. Canepari, G. Satta, and J. Coyette. 1980. Identification of the lethal target of benzylpenicillin in Streptococcus faecalis by in vivo penicillin binding studies. Nature 287:70-72. [DOI] [PubMed] [Google Scholar]
- 12.Fontana, R., R. Cerini, P. Longoni, A. Grossato, and P. Canepari. 1983. Identification of a streptococcal penicillin-binding protein that reacts very slowly with penicillin. J. Bacteriol. 155:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita, N., M. Yoshimura, T. Komori, K. Tanimoto, and Y. Ike. 1998. First report of the isolation of high-level vancomycin-resistant Enterococcus faecium from a patient in Japan. Antimicrob. Agents Chemother. 42:2150. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handwerger, S., D. C. Perlman, D. Altarac, and V. McAuliffe. 1992. Concomitant high-level vancomycin and penicillin resistance in clinical isolates of enterococci. Clin. Infect. Dis. 14:655-661. [DOI] [PubMed] [Google Scholar]
- 15.Kariyama, R., R. Mitsuhata, J. W. Chow, D. B. Clewell, and H. Kumon. 2000. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J. Clin. Microbiol. 38:3092-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 17.Lederberg, J., and E. M. Lederberg. 1951. Replica planting and indirect selection of bacterial mutants. J. Bacteriol. 63:399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ligozzi, M., F. Pittaluga, and R. Fontana. 1993. Identification of a genetic element (psr) which negatively controls expression of Enterococcus hirae penicillin-binding protein 5. J. Bacteriol. 175:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ligozzi, M., F. Pittaluga, and R. Fontana. 1996. Modification of penicillin-binding protein 5 associated with high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 40:354-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 21.Massidda, O., O. Dardenne, M. B. Whalen, W. Zorzi, J. Coyette, G. D. Shockman, and L. Daneo-Moore. 1998. The PBP 5 synthesis repressor (psr) gene of Enterococcus hirae ATCC 9790 is substantially longer than previously reported. FEMS Microbiol. Lett. 166:355-360. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto, T., T. Muratani, K. Okuda, M. Shiraisi, T. Hyashida, T. Oki, and M. Odawara. 2004. No resional spread of vancomycin-resistant enterococci with vanA or vanB in Kitakyusyu, Japan. J. Infect. Chemother. 10:331-334. [DOI] [PubMed] [Google Scholar]
- 23.Mazzulli, T., S. M. King, and S. E. Richardson. 1992. Bacteremia due to beta-lactamase-producing Enterococcus faecalis with high-level resistance to gentamicin in a child with Wiskott-Aldrich syndrome. Clin. Infect. Dis. 14:780-781. [DOI] [PubMed] [Google Scholar]
- 24.Murray, B. E. 1992. Beta-lactamase-producing enterococci. Antimicrob. Agents Chemother. 36:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray, B. E. 2000. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 342:710-721. [DOI] [PubMed] [Google Scholar]
- 26.Murray, B. E., and B. Mederski-Samaroj. 1983. A new mechanism for in vitro penicillin resistance in Streptococcus faecalis. J. Clin. Investig. 72:1168-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray, B. E., D. A. Church, A. Wanger, K. Zscheck, M. E. Levison, M. J. Ingerman, E. Aburutyn, and B. Mederski-Samoraj. 1986. Comparison of two beta-lactamase producing strains of Streptococcus faecalis. Antimicrob. Agents Chemother. 30:861-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray, B. E., K. V. Singh, S. M. Markowitz, H. A. Lopardo, J. E. Patterson, M. J. Zervos, E. Rubeglio, G. M. Eliopoulos, L. B. Rice, F. W. Goldstein, et al. 1991. Evidence for clonal spread of a single strain of beta-lactamase-producing Enterococcus faecalis to six hospitals in five states. J. Infect. Dis. 163:780-785. [DOI] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 1993. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 30.National Committee for Clinical Laboratory Standards. 2002. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 31.Osterblad, M., T. Leistevuo, and P. Huovinen. 1995. Screening for antimicrobial resistance in fecal samples by the replica planting method. J. Clin. Microbiol. 33:3146-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raze, D., O. Dardenne, S. Hallut, M. Martinez-Bueno, J. Coyette, and J. M. Ghuysen. 1998. The gene encoding the low-affinity penicillin-binding protein 3r in Enterococcus hirae S185R is borne on a plasmid carrying other antibiotic resistance determinants. Antimicrob. Agents Chemother. 42:534-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Signoretto, C., and P. Canepari. 2000. Paradoxical effect of inserting, in Enterococcus faecalis penicillin-binding protein 5, an amino acid box responsible for low affinity for penicillin in Enterococcus faecium. Arch. Microbiol. 173:213-219. [DOI] [PubMed] [Google Scholar]
- 35.Spratt, B. G., and A. B. Pardee. 1975. Penicillin-binding proteins and cell shape in Escherichia coli. Nature 254:516-517. [DOI] [PubMed] [Google Scholar]
- 36.Uttley, A. H., C. H. Collins, J. Naidoo, and R. C. George. 1988. Vancomycin-resistant enterococci. Lancet i:57-58. [DOI] [PubMed] [Google Scholar]
- 37.Williamson, R., C. Le Bouquenec, L. Gutmann, and T. Horaud. 1985. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J. Gen. Microbiol. 131:1933-1940. [DOI] [PubMed] [Google Scholar]
- 38.Williamson, R., S. B. Calderwood, R. C. Moellering, Jr., and A. Tomasz. 1983. Studies on the mechanism of intrinsic resistance to beta-lactam antibiotics in group D streptococci. J. Gen. Microbiol. 129:813-822. [DOI] [PubMed] [Google Scholar]
- 39.Witte, W., and I. Klare. 1995. Glycopeptide-resistant Enterococcus faecium outside hospitals: a commentary. Microb. Drug Resist. 3:259-263. [DOI] [PubMed] [Google Scholar]
- 40.Zhao, G., T. I. Meier, S. D. Kahl, K. R. Gee, and L. C. Blaszczak. 1999. Bocillin FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43:1124-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zighelboim-daun, S., and R. C. Moellering. Jr. 1988. Mechanisms and significance of antimicrobial resistance in enterococci, p. 610-613. In P. Actor, L. Daneo-Moore, M. L. Higgins, M. R. J. Salton, and G. D. Shockman (ed.), Antibiotic inhibition of bacterial cell surface assembly and function. American Society for Microbiology, Washington, D.C.
- 42.Zorzi, W., X. Y. Zhou, O. Dardenne, J. Lamotte, D. Raze, J. Pierre, L. Gutmann, and J. Coyette. 1996. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J. Bacteriol. 178:4948-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]