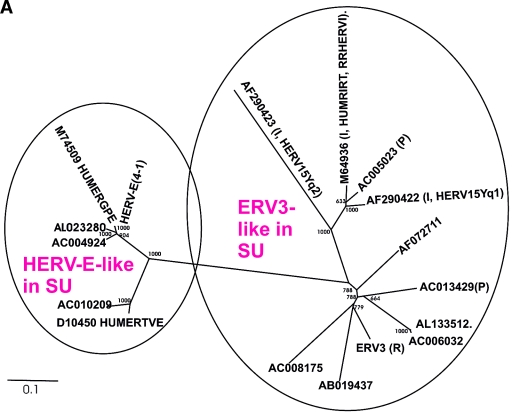
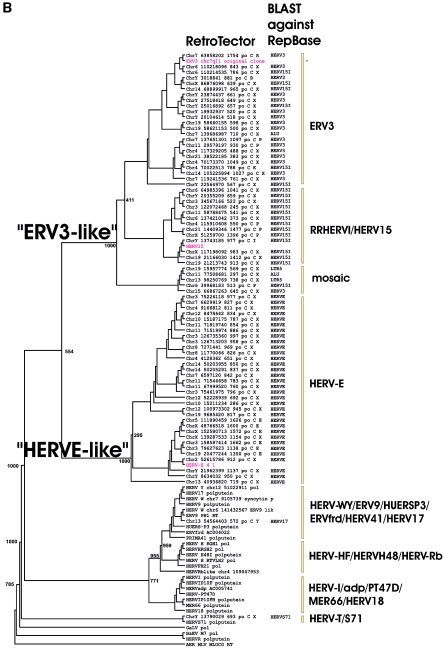
FIG. 2.
(A) Dendrogram of ERV3- and HERV-E-like sequences based on the nucleotide alignment of the SU part of env, partly shown in Fig. 3. PBS were analyzed by using the RetroTector program (see Materials and Methods) (G. O. Sperber and J. Blomberg, unpublished data). Sequences with an identifiable PBS are shown in parentheses, giving the amino acid (using the one-letter code) corresponding to parts of the tRNA sequence. (B) Pol amino acid-based cladogram, derived from the alignment shown in Fig. S3a (see the supplemental material). The neighbor-joining method was used. For each element, the following RetroTector-derived data are shown: chromosomal position, chain score, PBS usage, sum of stops and frame shifts, and the most similar RepBase element. The bootstrap value (1,000 replicates) for the branch which divides the ERV3-like and the HERVH HERV-E-like elements is also shown.