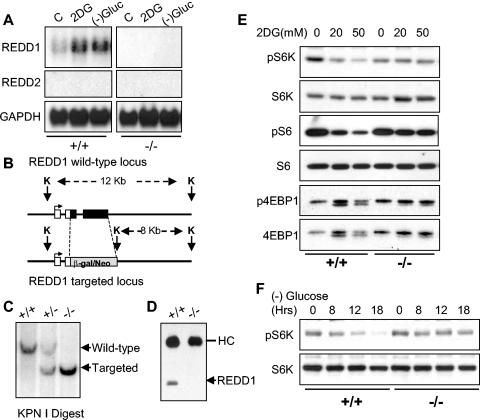
FIG. 1.
REDD1 is induced following energy stress and is required for mTOR substrate dephosphorylation. (A) Induction of the REDD1 message. Immortal MEFs of the indicated genotypes were washed, followed by addition of control medium with 10% fetal calf serum (C), the same medium/fetal calf serum with 2DG (25 mM), or glucose-free medium with 10% dialyzed fetal calf serum. Cells were harvested for Northern analysis at 4 h and probed with a REDD1 or REDD2 cDNA fragment. REDD2 expression is not detectable in MEFs, and no REDD1 expression is observed in REDD1−/− MEFs. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) REDD1 wild-type and targeted genomic alleles. White boxes, REDD1 3′ UTR; black boxes, REDD1 coding region and 5′ UTR; grey box, β-galactosidase/neo fusion cDNA. The sizes of KpnI (K) fragments detected in panel C are shown. (C) Southern blot analysis of KpnI-digested MEF DNA of the indicated genotype, probed with a genomic fragment 3′ of the REDD1 targeting construct (not shown). (D) Absence of REDD1 protein in REDD1−/− MEFs. Extracts from cells treated with 2DG to induce REDD1 protein were analyzed by immunoprecipitation-Western analysis using affinity-purified polyclonal REDD1 antisera. The immunoglobulin heavy chain (HC) is shown as a control for immunoprecipitation. (E) Dephosphorylation of S6K (T389), S6 (S235/236), and 4EBP1 (T70) is defective in REDD1−/− MEFs following ATP depletion. Western blot analysis of lysates from primary MEFs treated with 2DG for 4 h, probed with phosphospecific antibodies. The same blots were stripped and reprobed to determine the respective total protein levels. (F) S6K dephosphorylation (T389) in response to glucose starvation requires REDD1. Glucose withdrawal in MEFs was performed as in panel A for the indicated times prior to harvest for Western blot analysis as above.