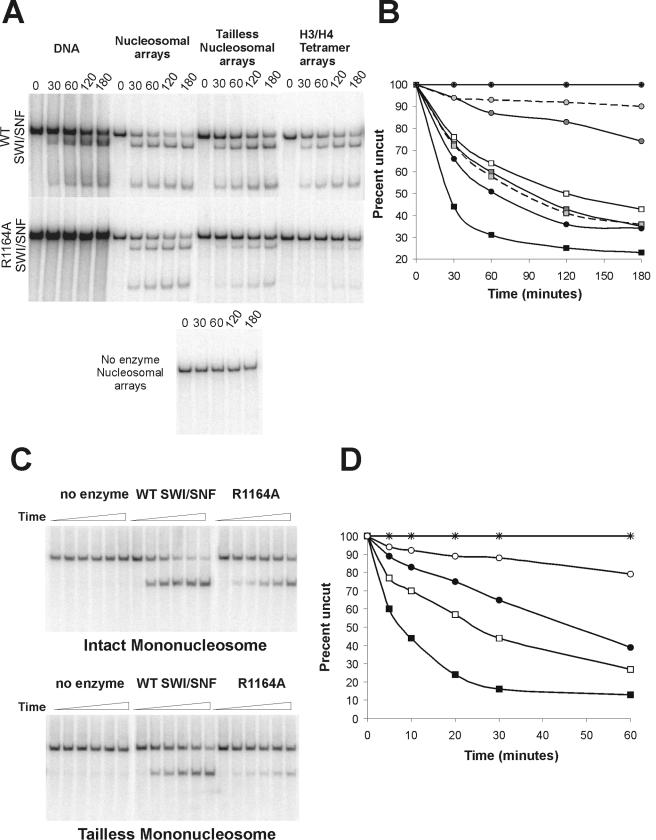
FIG.6.
Assembly of intact nucleosomes alleviates the remodeling defects of the R1164A SWI/SNF complex. (A) A 1.5 nM concentration of SWI/SNF was incubated with 0.1 nM nucleosomal cruciform template or 0.1 nM DNA template, 0.15 μg/ml endonuclease VII, and 3 mM ATP. (B) Graphical representation of data from panel A. The rates of torsion generation by no enzyme (⧫) or by wild-type (squares) or R1164A (circles) complexes were assayed on DNA (open symbols) or chromatin templates containing intact octamers (black symbols), tailless octamers (dark gray symbols), or (H3-H4)2 tetramers (light gray symbols connected by dashed lines). (C) The abilities of wild-type and R1164A SWI/SNF complexes to create restriction enzyme accessibility (HhaI) were tested on mononucleosomes with intact and tailless octamers. SWI/SNF (1.0 nM WT or R1164A) was incubated with 1 nM mononucleosomes, 3 mM ATP, and 40 U HhaI. Samples were removed from the reaction mixtures at specific time points over 1 h and prepared as described in Materials and Methods. (D) Graphical representation of data from panel C. Wild-type SWI/SNF (squares) or R1164A (circles) reactions were incubated with either intact octamers (filled symbols) or tailless octamers (open symbols). Reactions containing no enzyme were also incubated with intact octamers (×) or tailless octamers (+).