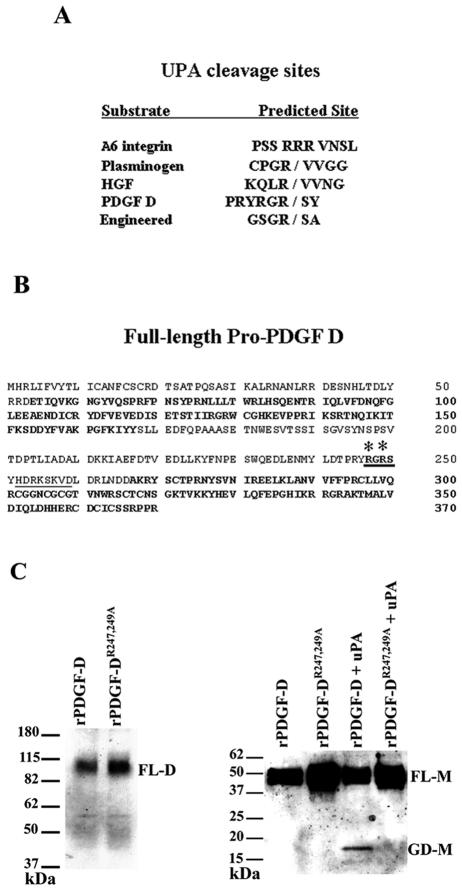
FIG. 7.
Mutational analysis identifies the uPA cleavage site in full-length PDGF D. (A) Comparison of uPA cleavage sites in engineered and natural uPA substrates. (B) Putative uPA cleavage site in the full-length PDGF D protein hinge region (cleavage site in bold and underlined). Asterisks indicate R247 and R249 mutated to alanines. The CUB domain and growth factor domain are in bold. The computer-predicted furin cleavage site is underlined. (C) Left panel, CV-1 cells were coinfected and transfected (see Materials and Methods) with vTF7-3 and either pTF7-PDGF DR247,249A or pTF7-PDGF D and cultured in serum-free medium for 24 h. CM was collected and resolved on an SDS-PAGE gel under nonreducing conditions to visualize the dimer form of full-length pro-PDGF D (∼80 kDa) using the anti-PDGF D-GD antibody. Right panel, rPDGF DR247,249A or rPDGF D was incubated at 37°C overnight with or without 10 nM uPA and then resolved on an SDS-PAGE gel under reducing conditions. PDGF D full-length (∼50 kDa) and growth domain (∼18 kDa) monomers were detected through immunoblot analysis using anti-PDGF D-GD antibody. FL-D, full-length PDGF D protein dimer; FL-M, full-length PDGF D protein monomer; GD-M, PDGF D growth domain monomer.