Figure 6.
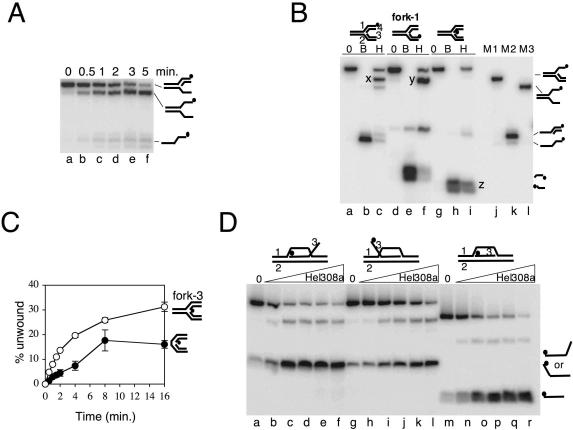
Hel308a unwinds lagging strands from nicks and the fork branchpoint. (A) Products from unwinding fork-2 by hel308a as a function of time. Reactions contained 2 nM DNA and 20 nM Hel308a in 5 mM MgCl2, 5 mM ATP at 45°C. (B) Reactions showing unwinding of strands in fork-1 by Hel308a. Cartoons of the substrate are shown above the panel in each case with the labelled strand denoted by a filled circle. Strands are numbered on one of these cartoons. Reactions were at 45°C for 20 min containing 5 mM MgCl2, 5 mM ATP, 2 nM DNA and zero (0) or 50 nM Hel308a (H). Lanes marked B contained no Hel308a and the reactions were heated to 95°C for 20 min. Letters X, Y and Z highlight the major product of unwinding in each reaction containing Hel308a. Substrate markers corresponding to fork lacking a lagging strand (M1), flayed duplex (M2) and partial duplex (M3) are annotated beside the panel. (C) Time-course unwinding of fork-3 and its corresponding nicked duplex DNA substrate. Substrates used are annotated to the right of the graph. Reactions were at 45°C for the times shown and contained 2 nM DNA, 5 mM MgCl2, 5 mM ATP and 20 nM Hel308a. Error bars derive from means of two independent assays. (D) Unwinding of the invading strand of D-loop substrates by Hel308a. The D-loop substrates used are annotated above the panel and reactions products are displayed to the right of the panel. Reactions were for 20 min at 45°C containing 2 nM DNA, 5 mM MgCl2, 5 mM ATP and zero or 1, 5, 10, 25 and 50 nM Hel308a.