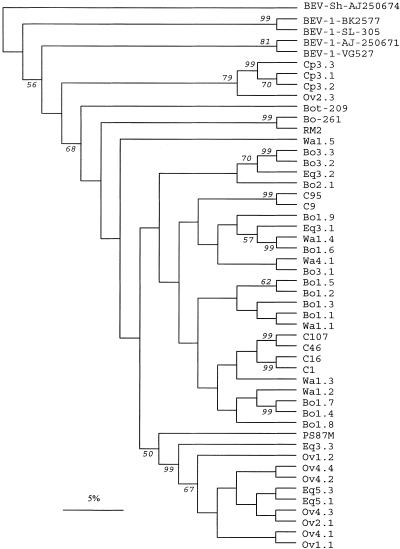
FIG. 3.
Phylogenetic tree of the amplified BEV 5′NCR of isolates used in the present study. Previously published BEV sequences from cattle (C) feces obtained in the United States (14), BEV-1 isolates SL305 (GenBank accession number AF123433), BK-2577 (AF123433), and VG-527 (D00214), a BEV-1 prototype (AJ250671), and BEV-2 isolates Bot-209 (AJ250673), Bo261 (AJ250672), RM2 (X79383), and PS87 (X79368), as well as the only available sheep sequence (AJ250674), were included in the tree for comparison. The designations for cattle (beginning with Bo), sheep (Ov), goat (Cp), horse (Eq), and water (Wa) samples used in the present study include two digits; the first digit indicates the sampling area (areas 1 to 5 in Fig. 1), and the second digit indicates the sample number. The numbers at the nodes indicate the bootstrap values (expressed as percentages) that support the adjacent nodes based on 100 resampling iterations. Only bootstrap values higher than 50% are shown. Scale bar = 5% nucleotide sequence divergence.