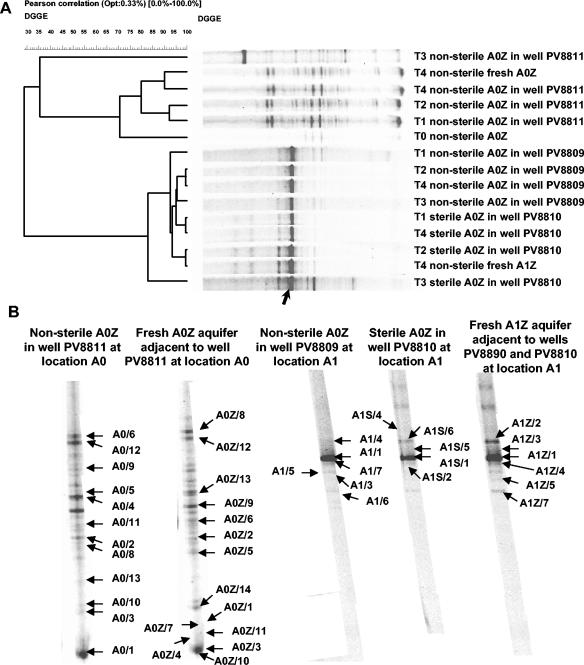
FIG. 3.
(A) UPGMA clustering of eubacterial 16S rRNA gene community fingerprints obtained from mesocosm aquifer material and fresh aquifer material at different times. Clustering was based on the Pearson moment-based similarity coefficient. T0, 9 January 2003; T1, 16 January 2003; T2, 30 January 2003; T3, 10 March 2003; T4, 20 May 2003. PV8809 is the contaminated well at location A1 into which microcosms containing nonsterile uncontaminated aquifer material were placed. PV8810 is the contaminated well at location A1 into which microcosms containing sterile uncontaminated aquifer material were placed. PV8811 is the well located in the uncontaminated area A0 into which microcosms containing nonsterile uncontaminated aquifer material were placed. The arrow indicates the dominant 16S rRNA gene band whose DNA showed 99% identity with the 16S rRNA gene sequence of Pseudomonas sp. strain SE22#1a. (B) Eubacterial 16S rRNA gene community fingerprints for samples obtained on 20 May 2003, with the cloned bands indicated (arrows). Based on BLASTN analysis, the 16S rRNA gene sequences corresponding to the indicated bands matched best with other 16S rRNA gene sequences as follows (accession numbers with the best match are given in parentheses): A0/1, Rhodococcus fascians ATCC 12974T (X81930); A0/2, Acidovorax sp. strain BSB421 (Y18617); A03, Rhodococcus sp. strain 5/14 (AF181690); A0/4, Rhodococcus sp. strain KU18 (AJ517832); A0/5, uncultured β-bacterium clone KD5-43 (AY218738); A0/6, Pseudomonas umsongensis Ps 3-10 (AF468450); A0/8, Pseudomonas sp. strain MSB2071 (AY275482); A0/9, Pseudomonas lanceolata ATCC 14669T (AB021390); A0/10, Arthrobacter sp. strain CS16 (AY371258); A0/11, Zoogloea sp. strain BAL15 (U63941); A0/12, Pseudomonas sp. strain MWH1 (AJ556801); A0/13, Rhodococcus sp. strain P_wp0233 (AY188941); A0Z/1, Rhodococcus fascians ATCC 12974T (X81930); A0Z/2, Bradyrhizobium sp. strain Hambi 2145 (AJ132567); A0Z/3, Rhodococcus erythropolis DCL14 (AJ131637); A0Z/4, Rhodococcus fascians ATCC 12974T (X81930); A0Z/5, Arthrobacter oxydans CF-46 (AJ243423); A0Z/6, uncultured beta-proteobacterium clone NMS8.125WL (AY043788); A0Z/7, Rhodococcus sp.strain P_wp0233 (AY188941); A0Z/9, uncultured beta-proteobacterium clone C47.33PG (AF431314); A0Z/10, Rhodococcus sp. strain P_wp0233 (AY188941); A0Z/11, Acidovorax sp. strain G8B1 (AJ012071); A0Z/12, Pseudomonas sp. strain MWH1 (AJ556801); A0Z/13, Rhodococcus sp. strain KU18 (AJ517832); A0Z/14, Arthrobacter sp. strain CS16 (AY371258); A1/1, Pseudomonas sp. strain SE22#1a (AY263477); A1/3, Rhodococcus sp. strain Lgg15.6 (AJ489362); A1/4, Pseudomonas fluorescens DSM50108 (D86002); A1/5, Pseudomonas sp. strain GOBB3-103 (AF321030); A1/6, Zoogloea sp. strain BAL15 (U63941); A1/7, Pseudomonas sp. strain NAF88 (AJ271413); A1S/1, Pseudomonas sp. strain SE22#1a (AY263477); A1S/2, Pseudomonas sp. strain NAF88 (AJ271413); A1S/4, Pseudomonas stutzeri API-2-142 (AJ410871); A1S/5, uncultured bacterium clone ZZ14C11 (AY214196); A1S/6, Pseudomonas fluorescens DSM50108 (D86002); A1Z/1, Pseudomonas sp. strain SE22#1a (AY263477); A1Z/2, Pseudomonas fluorescens DSM50108 (D86002); A1Z/3, uncultured bacterium clone ZZ14C11 (AY214196); A1Z/4, Pseudomonas sp. strain NAF88 (AJ271413); A1Z/5, Pseudomonas sp. strain MSB2074 (AY275484); A1Z/7, Pseudomonas sp. strain QSSC1-9 (AF170731).