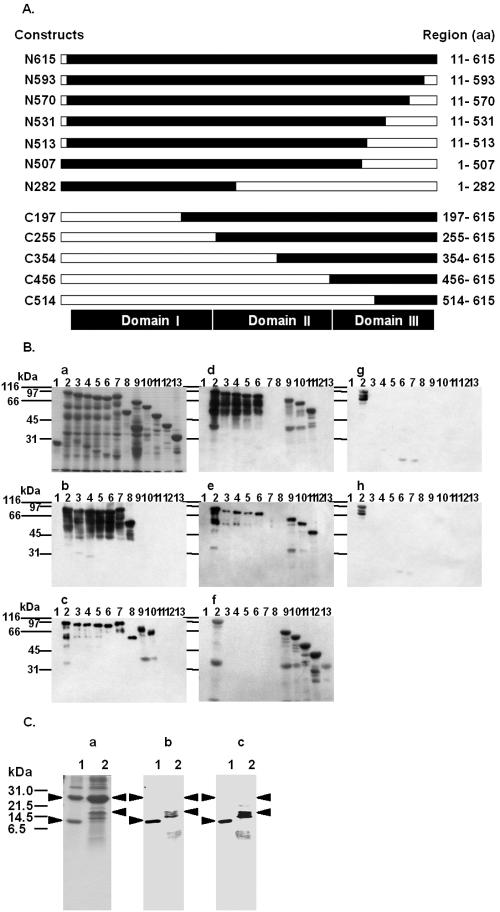
FIG. 5.
Immunoblot analysis of the reactivity of each MAb with Cry1Aa toxin deletion mutants. (A) Schematic diagram of Cry1Aa toxin deletion mutants. The open bars indicate the deleted portions of the full-length Cry1Aa toxin. The designation of each deletion mutant is indicated on the left, and the expressed regions are indicated on the right. aa, amino acids. (B) GST (lane 1) and approximately 2 μg of each of the 12 GST-fused Cry1Aa toxin deletion mutants, N615 (lane 2), N593 (lane 3), N570 (lane 4), N531 (lane 5), N513 (lane 6), N507 (lane 7), N282 (lane 8), C197 (lane 9), C255 (lane 10), C354 (lane 11), C456 (lane 12), and C514 (lane 13), were subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and probed with MAbs 3C7 (blot b), 1G10 (blot c), 1B10 (blot d), 2C2 (blot e), 1E10 (blot f), 2F9 (blot g), and 2A11 (blot h). The proteins on the blots were visualized by Coomassie brilliant blue staining (gel a). (C) Two Cry1Aa toxin C-terminal deletion mutants, C514 and C456, were expressed in E. coli at 25°C for 12 h. GST fusion proteins were purified from bacterial lysates by using glutathione-Sepharose 4B. Fusion proteins containing a thrombin recognition site were cleaved while they were bound to glutathione-Sepharose 4B. Approximately 1 μg of C514 (lane 1) and C456 (lane 2) in solution after enzymatic treatment was subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and probed with MAbs 1B10 (blot b) and 2C2 (blot c). The proteins on the gels were visualized by Coomassie brilliant blue staining (gel a). The arrowheads indicate the positions of the 26-kDa glutathione S-transferase (gel a, lanes 1 and 2), the 11-kDa thrombin fragment of C514 protein (lane 1), and the 17-kDa thrombin fragment of C456 (lane 2).