Abstract
Human papillomavirus (HPV) infection is the major cause of cervical cancer and its precursor, cervical intraepithelial neoplasia (CIN), and HPV testing has therefore been proposed for improved triaging and follow-up of women treated for CIN. We compared two common HPV DNA detection tests (Hybrid Capture II [HCII] and PCR-enzyme immunosorbent assay (EIA) using the primers GP5+/GP6+ followed by HPV typing with reverse dot blot hybridization) for sensitivity and specificity for detection of CIN and of CIN recurrence after treatment. Two hundred and thirty-nine women referred to the Department of Obstetrics and Gynaecology in Västerås, Sweden, were enrolled because of atypical Pap smears; 177 of these were later treated for dysplasia by conization or loop diathermy. Samples for HPV DNA testing were taken before and 4 to 6 months after treatment. There was substantial agreement between the HCII and PCR-EIA (kappa, 0.70 before treatment and 0.72 after treatment). The sensitivity for histopathologically confirmed CIN III was 100.0% for PCR-EIA and 95.6% for HCII. For patients with CIN II or worse (CIN II+), the sensitivities were 92.9% (PCR-EIA) and 91.8% (HCII). The specificities for CIN II+ in the pretreatment setting were 30.4% for PCR-EIA and 24.1% for HCII. After treatment, the sensitivities for CIN III in cytology were 100.0% by both methods, and for CIN II+, sensitivities were 80.0% by both methods. The specificities for CIN II+ in the posttreatment setting were 83.5% for PCR and 85.4% for HCII. In conclusion, the sensitivities of both PCR-EIA and HCII are high and almost equal, suggesting that both methods are suitable as tools for detection and posttreatment follow-up of CIN II-III.
Cancer of the cervix is the third most common form of cancer among women worldwide, with almost 400,000 new cases each year (28). The principal cause of invasive cervical cancer and its precursor lesions is infection with oncogenic types of human papillomavirus (HPV) (3, 4, 15, 21, 24, 34, 40), which is found in close to 100% of cancers (39). A recent case control study reported that HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82 conferred increased risk for cervical cancer and that HPV types 6, 11, 40, 42, 43, 44, 54, 61, 70, 72, and 81 did not increase the cervical cancer risk (23). A meta analysis of all studies of cervical cancer found that the most important HPV type in squamous cell carcinomas is HPV 16, followed by HPV 18, 45, 31, and 33 (7). In adenocarcinomas, the most common type is HPV 18 followed by HPV 16 and 45 (7).
In young women, the incidence of HPV infection is high, but the infection is often of short duration (16). The prevalence of high-risk (HR) HPV DNA is highest among sexually active teenagers, declining in women 20 to 30 years of age, and is still lower in women over the age of 30 (4, 19). Persistent HR HPV infections, which are more common in older women, greatly increase the risk of cervical intraepithelial neoplasia (CIN) (33) or cervical cancer, especially if the viral load is high (15, 16, 32). Regressing cervical lesions may clear their HPV some 3 months before cytological regression (25).
HPV testing is of interest for improvement of cervical cancer screening programs. If no HR HPV DNA is present in a cervical sample, it will be quite unlikely that the patient will develop cancer for some years (36). Secondary HPV testing of women with unclear or low-grade cytological abnormalities can therefore improve the specificity of cervical screening. Another important use of HPV testing is in the follow-up of patients treated for CIN. A positive HR HPV DNA test 6 months after treatment of CIN II/CIN III is more predictive for recurrence than an abnormal cytology (26). The most commonly used HPV DNA detection methods are PCR, using the general primer pairs MY09/11 and their derivatives PGMY09/11 (14) or GP5+/GP6+, and the Hybrid Capture II (HCII) test (Digene), a commercially available test and the second generation of this method. The second version is more accurate than the first version of Hybrid Capture, mainly because of a change of reagents and the addition of four new HR HPV types in the probe cocktail (31). In this study, we compared HCII and PCR with the general primer pair GP5+/GP6+ (9, 18) for detection of high-grade CIN among women referred for colposcopy as well as for detection of CIN recurrence after treatment. Some methodological comparisons between PCR and HCII have been published in the past (35, 37), but we have not found any studies that have focused on comparing clinical sensitivity and specificity in detection and posttreatment follow-up of high-grade CIN.
MATERIALS AND METHODS
Study design.
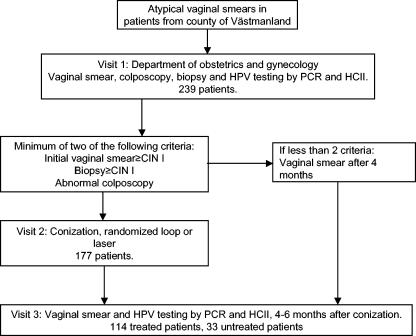
The present study contains 239 women living in the county of Västmanland, Sweden, who attended organized population-based cervical cancer screening and had been referred to the Department of Obstetrics and Gynaecology in Västerås because of atypical smears. All referred women were eligible for inclusion. The cytological diagnosis of the smear that had resulted in referral was available for 197 women: 41% had had cytological findings of unclear significance, 1% had “unclear atypia,” 35.5% had CIN I, 14.2% had CIN II, and 8.1% had CIN III. At the first visit, an endo/ectocervical sample was taken using a cytobrush (Medscand, Malmö, Sweden) and used for cytology (Fig. 1). For the present study, the same brush was thereafter immersed in 1 ml 0.9% NaCl, vigorously stirred, and frozen at −20°C. A second cervical sample was taken using the brush and sample transport medium provided with the HCII “sample collection kit,” all according to the instructions of the manufacturer.
FIG. 1.
Study flow chart.
The taking of a conventional smear before the taking of the HCII sample has not been reported to affect the performance of the HCII test; e.g., in a recent cohort study, 1,278 women had a smear taken before the HCII sample was taken, and 3,123 gave an HCII sample without first taking a smear. The HCII test had excellent performance in the entire cohort (99.9% negative predictive value) (6).
A colposcopy-directed biopsy was taken for routine histopathology. In order to be referred to the second visit, at least 2/3 of the following criteria had to be fulfilled: the initial Pap smear and/or biopsy showed CIN I or more and the colposcopy showed pathological signs. At the second visit, depending on the result of the biopsy, a conization was performed with either loop or laser (choice of method was allocated at random).
All conization specimens were reviewed by the same pathologist. In case of disagreement with the routine histopathological diagnosis, the histopathological diagnosis of the review was used.
At the third visit 4 to 6 months after conization, samples for cytology and HPV testing with PCR-enzyme immunosorbent assay (EIA) and HCII were obtained. Women who were not treated were also asked for a follow-up cervical sample 4 to 6 months after colposcopy. When the present work was closed, 239 women had been enrolled and had attended visit 1; 177 of these had been treated. A total of 147 women had attended the first follow-up visit; 114 of these had been treated. Two women did not attend the first follow-up visit because of hysterectomy, and one didn't attend because of relocation. For a further 89 patients that were enrolled shortly before the present part of the study was closed, no follow-up visits had as yet been scheduled, and they are therefore not included in this report.
The study protocol was approved by the institutional review board of Västerås Hospital.
PCR-EIA method.
The samples, which had the initial volume of approximately 1,000 μl, were thawed and centrifuged at 3,000 × g for 10 min at room temperature. The cell pellet was resuspended in 1,000 μl Tris-HCl (10 mM), pH 7.4. Aliquots of each sample were frozen at −20°C, thawed and boiled for 10 min, centrifuged briefly to avoid droplets at the tube top, and frozen at −20°C until analysis was performed.
Sample preparation and PCR using the general primer pair GP5+/GP6+ (18) were performed in separate rooms. Volumes of 10 μl from each sample were added to a master mix with 0.5 μM of the primer GP5+ and the biotinylated primer GP6+ (supplied from PJF Snijders, Free University of Amsterdam, Holland), 3.5 mM MgCl2, 200 μM of each deoxynucleoside triphosphate, 1 U of AmpliTaq Gold DNA polymerase, and PCR buffer II (Roche) to a final volume of 50 μl. As positive controls of the PCR system, 10-fold dilutions (10 ng to 100 pg) of purified HPV 16 DNA from SiHa cells in a background of 100 ng of COT-1 human placental DNA (Roche, Germany) were analyzed in all tests. The extraction of positive controls was performed with a sodium dodecyl sulfate (SDS)-proteinase K method, as previously described (12), with some modifications. The volume of saturated ammonium acetate was 150 μl, the incubation at 37°C was performed overnight, and all centrifugations were performed at 16,000 × g. A serial endpoint dilution of purified HPV 16 plasmids (10-fold dilutions of 104 to 10−2 copies/μl) was used to determine assay sensitivity, which was found to be 10 copies (equivalent to 0.13 fg/reaction or 2.6 fg/ml) of HPV 16 DNA per sample. The samples were tested for amplifiability in a separate PCR with primers for the human β-globin gene, BGPCO5 and biotinylated BGPCO3 (DNA Technology, Denmark), and AmpliTaq DNA polymerase. The positive controls of this system were 10 ng and 1 ng of human placental DNA (Sigma, Germany). As negative controls, 10 μl of sterile water was applied to separate reaction tubes in the absence of template and analyzed identically as the other samples. Water controls with water added both before and after the samples were included in all runs.
The PCR was performed in a Hybaid Omnigene automated thermal cycler (Hybaid, United Kingdom). For the PCR using the GP+ primers, a first step at 94°C for 10 min was followed by 45 cycles of denaturing at 94°C for 1.5 min, annealing at 40°C for 1.5 min, an extension step at 72°C for 2 min, and a terminal extension step at 72°C for 4 min. The PCR program using the primers for the human β-globin gene was initiated with a step at 94°C for 4 min followed by 40 cycles of 94°C for 1.5 min, 45°C for 1.5 min, 72°C for 2 min, and a final step at 72°C for 4 min.
For detection of amplified DNA, 5 μl of each biotinylated PCR product was applied to streptavidin-coated microtiter plate wells (Roche, Germany), together with 50 μl of 1× SSC (0.15 M NaCl and 15 mM sodium citrate, pH 7.0) supplemented with 0.5% Tween 20 (Sigma, Germany). The plate was covered and incubated for 60 min at 37°C and then washed with 3 × 200 μl of 1× SSC-0.5% Tween 20. For denaturation, 0.2 M NaOH was added followed by incubation for 15 min at room temperature. The previously described washing was repeated, and probe solution containing 10 nM of each probe diluted in 1× SSC-0.5% Tween 20 was added to each well, after which the plate was incubated for 60 min at 37°C. After new washings, alkaline phosphatase-conjugated anti-digoxigenin antibody (75 mU/ml; Roche, Germany) diluted in 1× SSC-0.5% Tween 20 was added, and the plate was incubated as before. Finally, the plate was washed with 5 × 200 μl of 1× SSC-0.5% Tween 20, and 100 μl of alkaline phosphatase substrate (Fast p-Nitrophenyl Phosphate tablet in sterile water; Sigma, Germany) was added to each well. The optical density (OD) was measured at 405 nm after an overnight incubation at 37°C. The probe solution for the GP5+/GP6+ PCR product consists of 14 different oligonucleotide probes for the HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68 (supplied from P. J. F. Snijders, Holland), and each probe is labeled with digoxigenin-11-ddUTP (18). For the β-globin gene PCR products, the same procedure as above was repeated except for the use of a single, digoxigenin-labeled probe diluted in 1× SSC-0.5% Tween 20 (5′-AAG AGT CAG GTG CAC CAT GGT GTC TGT TTG-3′; DNA Technology, Denmark).
For a valid GP5+/GP6+ PCR-EIA, the positive control with 100 pg HPV 16 DNA had to be positive. The cutoff was three times the mean OD value of negative controls after overnight incubation. The “grey zone” was set to between two and three times the mean OD value of the negative controls. All samples that had OD values above cutoff or in the grey zone were tested in reverse dot blot hybridization (RDBH) (13) and scored as positive if an HPV type could be identified. For the β-globin PCR-EIA, the positive control containing 10 ng of human placental DNA had to be positive. The cutoff was set as it was for HPV.
Type-specific PCR method.
Type-specific PCRs for HPV 16 and 18 were performed in a Mastercycler (Eppendorf). A first step at 94°C for 3 min was followed by 40 cycles of denaturing at 94°C for 1.5 min, annealing at 50°C for 1.5 min, and an extension step at 72°C for 1.5 min. The following primers target the E6 gene and are for HPV 16: 31-50, biotinylated at the 5′ end (5′-CG TAA CCG AAA TCG GTT GAA-3′), and 123-106 (5′-TCC TGT GGG TCC TGA AAC-3′). For HPV 18, the primers were 54-72, biotinylated at the 5′ end (5′-CG GGA CCG AAA ACG GTG TA-3′), and 130-112 (5′-CGT GTT GGA TCC TCA AAG C-3′). The size of the amplicons are approximately 90 bp for HPV 16 and 80 bp for HPV 18.
A sample volume of 2.5 μl was added to a master mix consisting of 0.75 μM of each primer (DNA Technology, Denmark), 200 μM of each deoxynucleoside triphosphate, 0.2% bovine serum albumin, 1× Dynazyme buffer, and 0.625 U of Dynazyme II DNA polymerase (Finnzymes, Finland) to a final volume of 25 μl. As positive controls of the HPV 16 PCR, we analyzed 10-fold dilutions, 2.5 ng to 2.5 pg, of purified HPV 16 DNA from SiHa cells in a background of 25 ng of COT-1 human placental DNA (Roche, Germany). Tenfold dilutions (104 to 10−2 copies/μl) of HPV 16 plasmids have been analyzed in this PCR system, showing that it detects 2.5 copies, which is equivalent to 0.033 fg or 1.3 fg/ml, of HPV 16 plasmids per sample. Positive controls of the HPV 18 PCR were 10-fold dilutions, 3.3 ag to 3.3 fg, of HPV 18 DNA in pBR322 plasmids in a background of 25 ng human placental DNA, and this system detects 25 copies, equivalent to 0.33 fg or 13.3 fg/ml, of HPV 18 plasmids per sample. The negative controls were the same as those for the GP5+/6+ PCR.
The detection of the PCR products was performed by the EIA procedure, as described above, except that the probes used were 5′-AT TGC AGT TCT CTT TTG GTG CAT AAA ATG TC-3′ for HPV 16 and 5′-GC GCC ATA GTA TTG TGG TGT GTT TCT CAC AT-3′ for HPV 18 (DNA Technology, Denmark).
HPV typing with RDBH.
The HPV typing was performed as previously described (13), with some modifications.
Briefly, 100 ng of HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68 plasmids and 33 ng of β-globin DNA plasmid were denatured in sterile 0.8 M NaOH-50 mM EDTA and dot blotted onto Hybond N+ membranes (Amersham, United Kingdom) prewetted in 6× SSC (0.9 M NaCl plus 90 mM sodium citrate, pH 7.0). After drying at room temperature and a 20-min incubation at 120°C, the membranes were dipped in 2× SSC and incubated on nylon filters in 5 ml of 1 M NaCl-50% deionized formamide-20% dextran sulfate 50-1% SDS-0.2 mg herring sperm DNA for 60 min at 46°C. Five microliters (in case of “grey zone” samples, 10 μl) of the biotinylated PCR product from the GP5+/GP6+ PCR-EIA in 50 μl prehybridization solution was incubated for 5 min at 94°C. Two microliters of the biotinylated PCR product from the β-globin PCR-EIA was also added. Hybridization was performed overnight at 46°C. After washing with 2× SSPE-0.1% SDS (0.3 M NaCl, 20 mM NaH2PO4, and 2 mM EDTA, pH 7.4) for 15 min at 65°C, 3% bovine serum albumin-154 mM NaCl-50 mM Trizma base-0.05% Tween 20 (filtered through 0.45-μm filters) was added followed by incubation for 60 min at 65°C. Streptavidin-alkaline phosphatase conjugate (Invitrogen) diluted 1/3,333 in 154 mM NaCl-50 mM Trizma base-0.05% Tween 20 (filtered through 0.22-μm filters) (TBS-T) was incubated for 10 min at 20 to 40°C. After two further washings with TBS-T for 10 min at 20 to 40°C and washing in 100 mM Tris-HCl (pH 9.5)-100 mM NaCl-50 mM MgCl2, the membranes were briefly dried and incubated with Lumi-Phos 530 (Lumigen) for 75 min at room temperature before exposure for 10 min to X-ray film (Kodak) between intensifying screens in a cassette (Cronex Lightning Plus; DuPont).
HCII method.
The Hybrid Capture II test is a nucleic acid hybridization assay where specimens containing the target DNA hybridize with a specific HPV RNA probe mixture including probes for the following HR HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68, i.e., the same types as those detected in the GP5+/6+ PCR-EIA method with the exception of HPV 66. The resultant DNA:RNA hybrids are captured on a microplate coated with antibodies specific for DNA:RNA hybrids. After signal detection with antibodies conjugated with alkaline phosphatase and substrate, the emitted light is measured in a luminometer as relative light units (RLU). Samples are classified as positive for HR HPV if the relative light unit (RLU) reading is above 1.0, which (according to the manufacturer) is equivalent to 1 pg HPV DNA/ml. We did not test for low-risk HPV types. Borderline results (0.7 to 2.0 RLU) (found in 27 samples) were retested in duplicates, and a 2/3 decision on whether the specimen was positive or not was made.
RESULTS
Concordance pretreatment.
Before treatment, all 239 enrolled women were tested for HPV DNA, and 177 of these women were then treated by conization at visit 2. The majority of all enrolled women, 162, were positive for HPV by both methods, 49 were negative by both methods, 18 were negative by PCR and positive by HCII, and 10 were positive by PCR and negative by HCII (kappa, 0.70).
In the subpopulation of the 177 treated women, 88 out of 98 patients with CIN II or worse (CIN II+) in histopathology were HPV positive by both methods (Table 1). Five patients were negative by both methods, seven were negative by PCR, and eight were negative by HCII (Table 1) (kappa, 0.639). Four of the five patients with CIN II that were negative by both methods were reanalyzed and again found negative by both methods. Among the 30 patients with normal histopathology (which includes the diagnosis “koilocytosis” in accordance with the nomenclature of Koss [20]), 23 patients were positive by HCII and 19 were positive by PCR, respectively.
TABLE 1.
HPV DNA test results from PCR and HCII analyses of samples before conization from the 177 patients that were treateda
| Sample type | No. of patients
|
||||
|---|---|---|---|---|---|
| PCR+/HC+ | PCR+/HC− | PCR−/HC+ | PCR−/HC− | Total | |
| Normal | 14 | 1 | 5 | 4 | 24 |
| Koilocytosis | 3 | 1 | 1 | 1 | 6 |
| CIN I | 34 | 2 | 3 | 10 | 49 |
| CIN II | 45 | 1 | 2 | 5 | 53 |
| CIN III/CIS | 43 | 2 | 0 | 0 | 45 |
The results are related to the histopathological diagnosis at treatment. CIS, carcinoma in situ.
Sensitivity, specificity, and predictive values for CIN II-III by histopathology and cytology.
The performance indicators of HCII and PCR-EIA were quite similar, both with and without reanalysis of discrepant samples (summarized in Table 2).
TABLE 2.
Sensitivities, specificities, positive predictive values, and negative predictive values of HCII and PCR with and without reanalysis of discrepant samples
| Clinical endpoint | Performance indicatora | HCII (%) | HCII with reanalysis (%) | PCR (%) | PCR with reanalysis (%) |
|---|---|---|---|---|---|
| Histopathology, CIN II+ | Sensitivity | 91.8 | 90.8 | 92.9 | 92.9 |
| Specificity | 24.1 | 30.4 | 30.4 | 27.8 | |
| PPV | 60.0 | 61.8 | 62.3 | 61.5 | |
| NPV | 70.4 | 72.7 | 77.4 | 75.9 | |
| Histopathology, CIN III | Sensitivity | 95.6 | 95.6 | 100.0 | 100.0 |
| Specificity | 18.9 | 23.5 | 23.5 | 22.7 | |
| PPV | 28.7 | 29.9 | 30.8 | 30.6 | |
| NPV | 92.6 | 93.9 | 100.0 | 100.0 | |
| Cytology, CIN II+ | Sensitivity | 80.0 | 80.0 | 80.0 | 80.0 |
| Specificity | 85.4 | 85.4 | 83.5 | 83.5 | |
| PPV | 21.1 | 21.1 | 19.0 | 19.0 | |
| NPV | 99.0 | 99.0 | 99.1 | 99.0 | |
| Cytology, CIN III | Sensitivity | 100.0 | 100.0 | 100.0 | 100.0 |
| Specificity | 84.0 | 84.0 | 82.1 | 82.1 | |
| PPV | 10.5 | 10.5 | 9.5 | 9.5 | |
| NPV | 100.0 | 100.0 | 100.0 | 100.0 |
PPV, positive predictive values; NPV, negative predictive values.
Concordance posttreatment.
After treatment (visit 3), 111/147 patients were negative by both methods, 23 patients were positive by both methods, 6 were negative by PCR and positive by HCII, and 7 were positive by PCR and negative by HCII (kappa, 0.72) (data not shown).
Most patients (57/68) with CIN II+ in histopathology at treatment became HPV negative by both methods after treatment (Table 3). Seven out of 68 CIN II+ patients were HPV positive by both methods also after treatment. Only four patients were discrepant, all being positive by PCR and negative by HCII (PCR+/HCII−) (kappa, 0.746). Among the 22 patients that had had normal histopathology, 6 were HPV positive by both methods after treatment (Table 3). Only two patients were discrepant (both negative by PCR and positive by HCII [PCR−/HCII+]).
TABLE 3.
HPV DNA test results from PCR and HCII analyses of samples from 114 patients after conizationa
| Sample type | No. of patients
|
||||
|---|---|---|---|---|---|
| PCR+/HC+ | PCR+/HC− | PCR−/HC+ | PCR−/HC− | Total | |
| Normal | 5 | 0 | 1 | 11 | 17 |
| Koilocytosis | 1 | 0 | 1 | 3 | 5 |
| CIN I | 4 | 2 | 1 | 17 | 24 |
| CIN II | 5 | 2 | 0 | 32 | 39 |
| CIN III/CISb | 2 | 2 | 0 | 25 | 29 |
The HPV DNA test results are related to the histopathological diagnosis of the cone at treatment.
CIS, carcinoma in situ.
Five out of 110 patients had CIN II+ in cytology at the first follow-up visit; four of these were HPV positive by both methods, and one was HPV negative by both methods (Table 4). Most patients, 83/110, had normal cytology and were HPV negative by both methods. Only 8 out of 110 patients had normal cytology and were HPV positive by both methods. There were six patients that were discrepant among patients with normal cytology: four were PCR+/HCII−, and two were PCR−/HCII+ (Table 4).
TABLE 4.
HPV DNA test results from PCR and HCII analyses of samples from 110 patients (all treated patients) related to the cytological diagnosis at the first follow-up visit
| Sample type | No. of patients
|
||||
|---|---|---|---|---|---|
| PCR+/HC+ | PCR+/HC− | PCR−/HC+ | PCR−/HC− | Totala | |
| Normal | 8 | 4 | 2 | 83 | 97 |
| CIN I | 4 | 1 | 1 | 0 | 6 |
| CIN II | 2 | 0 | 0 | 1 | 3 |
| CIN III/CISb | 2 | 0 | 0 | 0 | 2 |
| Cytological sample not adequate | 1 | 0 | 0 | 1 | 2 |
Another four patients should have been included, but it was not possible to obtain any adequate cytological samples from these patients.
CIS, carcinoma in situ.
Reanalysis of discrepant samples.
Forty-one samples had discrepant results in PCR-EIA and HCII (28 pretreatment and 13 posttreatment samples). All discrepant samples were analyzed again with PCR and HCII as well as with type-specific PCR (TS-PCR) for types 16 and 18. Two out of four HCII-negative samples that were HPV 16 positive by PCR-EIA/RDBH were negative in TS-PCR. However, the repeat PCR-EIA/RDBH did again find that these samples were HPV 16 positive. Similarly, one out of four HCII-negative samples that were HPV 18 positive by PCR-EIA/RDBH was negative in TS-PCR, but the repeat PCR-EIA/RDBH did again find this sample to be HPV 18 positive (not shown).
Among the 28 discrepant pretreatment samples, 8 initially HCII-positive samples, but none of the initially PCR-positive samples, became negative. None of the initially HCII-negative samples became positive, but two initially PCR-negative samples became positive. Thirteen posttreatment samples were discrepant. The only change on retesting was two HCII-positive samples that became negative. Among the 10 HCII-positive samples that became negative in reanalysis, 6 had had borderline results. For 27 out of the 41 samples that were reanalyzed by HCII, the relative light unit values decreased after each rerun (not shown).
Discrepant samples in relation to histopathology at treatment.
Most (13/18) patients with positive HCII and negative PCR pretreatment samples had normal histopathology or were not treated, 5/18 patients had CIN I-II, and none had CIN III. Similarly, most (5/6) patients with positive HCII and negative PCR posttreatment samples had normal histopathology at treatment or had not been treated, and only 1/6 had CIN I at treatment. Among patients with PCR+/HCII− pretreatment samples, 5/10 patients had normal histopathology or were untreated, 3/10 had CIN I-II, and 2/10 had CIN III. Among patients who were PCR+/HCII− at follow-up, most (6/7) had CIN before treatment (one patient had not been treated, four had CIN I-II, and two had CIN III).
DISCUSSION
We found a substantial concordance between PCR and HCII (kappa values of 0.70 and 0.72 before and after treatment, respectively), which is in accordance with previous studies (2, 30, 37). However, the number of positive samples detected by HCII tended to be higher than the number detected by PCR-EIA among patients with normal histopathology, both before treatment (HCII, 23 samples; PCR, 19 samples) and after treatment (HCII, 8 samples; PCR, 6 samples). One possible explanation for this could be cross-reactivity with other HPV types. Most HPV types previously found to cause false positivity by cross-reactivity with the probe cocktail in the HCII test are not classified as HR HPVs (1, 30, 35) and would be expected to be found preferentially in histopathologically normal specimens.
Several PCR-negative and HCII-positive samples (9/24; 38%) became PCR negative and HCII negative on reanalysis, particularly those which had had borderline HCII results (6/9; 67%). In the HCII reanalysis, the relative light unit values for 27/41 (66%) samples decreased. This could be explained either by the fact that only discrepant samples were selected for reanalysis (“regression dilution bias” [22]) or if the specimens were not stable on storage.
Four out of five samples from patients diagnosed with CIN II were reanalyzed and repeatedly HPV negative in both tests. Several reasons for this are possible. The samples may have been truly HPV negative. It is, e.g., reported that regressing CIN lesions may become HPV negative some time before they show morphological regression (25). Other possibilities include infection with an HR HPV type not included in the HCII and PCR-EIA probe cocktails or histopathological misdiagnosis.
The somewhat better sensitivity of PCR-EIA than HCII, 100% compared to 95.6%, respectively, for detection of CIN III is not due to differences in probe composition between PCR and HCII, since only one more HPV type (HPV 66) is included in the PCR probe cocktail, and none of the discrepant samples in the entire study was positive for HPV 66. PCR had a higher analytical sensitivity (2.6 fg of HPV 16 plasmids/ml) than HCII (1 pg HPV DNA/ml according to the manufacturer), and it is therefore likely that the PCR+/HCII− samples contained low amounts of virus. Notably, PCR+/HCII− samples were more common than PCR−/HCII+ samples in the follow-up samples taken after treatment, in line with the possibility that there are only few infected cells left after conization.
Improved follow-up after CIN treatment is a major application of HPV testing. Successful treatment for CIN is associated with HPV clearance (11). HPV DNA persistence is known to be a near-necessary risk factor for CIN (3, 4, 15, 21, 24, 34, 40), and as expected, posttreatment presence of HR-HPV is a good marker for recurrence (10, 17), better than abnormal cytology (26).
The recent systematic literature review of Paraskevaidis et al. (27) identified 11 studies (eight retrospective and three prospective studies) that evaluated the use of HPV testing after treatment for CIN. The total number of women included in these studies was 900, of whom 672 (75.3%) were considered as having a successful treatment, compared with 204 (23.3%) who were considered treatment failures.
Nine studies used PCR and two studies used HCII; none of the studies used both tests. The specificity of HPV testing ranged from 44% to 95%. Altogether, among the 672 women in whom the treatment was considered successful, the postoperative HPV DNA test was reported as negative in 566 (84.2%) women and positive in 106 (15.8%) women. In contrast, among the 204 cases that were considered as treatment failures, only 35 cases (17.2%) had a negative postoperative HPV DNA test, whereas 169 cases (82.8%) tested positive.
The results of the systematic review (27) are well in line with our study: 97 of our patients had normal cytology posttreatment, 10 of which were HPV positive by HCII (10.3%) and 12 of which were positive by PCR (12.4%). Six out of 110 treated patients (5%) had CIN I and 5/110 (4.5%) had CIN II+ in cytology after treatment (two patients had CIN III, and three had CIN II). For both HCII and PCR, 9/11 (82%) of the patients with CINI, CIN II, or CIN III in posttreatment cytology were HPV positive posttreatment. One of the patients with CIN II was negative by both PCR-EIA and HCII, whereas the other four with CIN II+ were positive by both methods, and there were no discrepant results. The negative predictive values for CIN II+ in cytology posttreatment were 100% for CIN III and 99% for CIN II+ for both methods, which confirms the value of HPV testing in follow-up after treatment, since a negative HPV DNA test posttreatment implies a negligible risk of residual high-grade disease (17). Our data suggest that the variability between various studies (27) in performance of HPV DNA testing in posttreatment follow-up is not due to the type of HPV test used and that both methods appear to be adequate for identifying patients that are not at high risk of residual or recurrent disease after treatment. Possible reasons for variability between studies include differences in laboratory performance or in study design.
Cervical screening using a combination of HPV DNA testing by HCII and cytology has substantially better sensitivity for CIN II+ detection than cytology alone, but the specificity is lower (2, 5, 29). The sensitivity of HCII alone for detecting CIN II+ lesions has been reported to be >95% (5, 8). We find similar sensitivities for CIN II+ detection by PCR-EIA and HCII, at levels comparable to those of previous reports (100.0% for CIN III and 92.9% for CIN II+ by PCR, and 95.6% for CIN III and 91.8% for CIN II+ by HCII). However, the specificities were low, 23.5% for CIN III and 30.4% for CIN II+ by PCR and 18.9% for CIN III and 24.1% for CIN II+ by HCII. It should be noted that the specificity refers to the specificity in a clinical secondary screening setting where all women have had an abnormal smear and that the specificities in a primary screening setting would most likely be substantially better. The HCII method is easy to use and is commercially available and has therefore been recommended for routine screening use (5, 8, 37, 38), although some authors suggest that the number of false-positive samples in normal smears due to cross-reactivity with low-risk HPVs needs to be reduced (35). We confirm that HCII had some false-positive samples among patients with normal histopathology, but the concordance between HCII and PCR was generally good, and the sensitivity of HCII for CIN III detection was high and almost equal to the sensitivity of PCR. In summary, both for CINII+ detection in a secondary screening setting as well as for follow-up posttreatment, both HCII and PCR appear to be adequate for routine use.
Acknowledgments
This study was supported by Europe against Cancer and by the Swedish Cancer Society.
We thank Kia Sjölin, University Hospital of Malmö, for technical assistance.
REFERENCES
- 1.Atypical Squamous Cells of Undetermined Significance/Low-Grade Squamous Intraepithelial Lesions Triage Study (ALTS) Group. 2000. Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. J. Natl. Cancer Inst. 92:397-402. [DOI] [PubMed] [Google Scholar]
- 2.Bergeron, C., D. Jeannel, J. D. Poveda, P. Cassonnet, and G. Orth. 2000. Human papillomavirus testing in women with mild cytologic atypia. Obstet. Gynecol. 95:821-827. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, and K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, F. X., and N. Munoz. 2002. The viral etiology of cervical cancer. Virus Res. 89:183-190. [DOI] [PubMed] [Google Scholar]
- 5.Clavel, C., M. Masure, J. P. Bory, I. Putaud, C. Mangeonjean, M. Lorenzato, R. Gabriel, C. Quereux, and P. Birembaut. 1999. Hybrid capture II-based human papillomavirus detection, a sensitive test to detect in routine high-grade cervical lesions: a preliminary study on 1518 women. Br. J. Cancer 80:1306-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clavel, C., J. Cucherousset, M. Lorenzato, S. Caudroy, J. M. Nou, P. Nazeyrollas, M. Polette, J.-P. Bory, R. Gabriel, C. Quereux, and P. Birembaut. 2004. Negative human papillomavirus testing in normal smears selects a population at low risk for developing high-grade cervical lesions. Br. J. Cancer 90:1803-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clifford, G. M., J. S. Smith, M. Plummer, N. Munoz, and S. Franceschi. 2003. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br. J. Cancer 88:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuzick, J., E. Beverley, L. Ho, G. Terry, H. Sapper, I. Mielzynska, A. Lorincz, W. K. Chan, T. Krausz, and P. Soutter. 1999. HPV testing in primary screening of older women. Br. J. Cancer 81:554-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Roda Husman, A.-M., J. M. M. Walboomers, A. J. C. van den Brule, C. J. L. M. Meijer, and P. J. F. Snijders. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 76:1057-1062. [DOI] [PubMed] [Google Scholar]
- 10.Distefano, A. L., M. A. Picconi, L. V. Alonio, D. Dalbert, J. Mural, O. Bartt, G. Bazan, G. Cervantes, M. Lizano, A. G. Carranca, and A. Teyssie. 1998. Persistence of human papillomavirus DNA in cervical lesions after treatment with diathermic large loop excision. Infect. Dis. Obstet. Gynecol. 6:214-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elfgren, K., P. Bistoletti, L. Dillner, J. M. M. Walboomers, J. L. M. Meijer, and J. Dillner. 1996. Conization for cervical intraepithelial neoplasia is followed by disappearance of human papillomavirus deoxyribonucleic acid and a decline in serum and cervical mucus antibodies against human papillomavirus antigens. Am. J. Obstet. Gynecol. 174:937-942. [DOI] [PubMed] [Google Scholar]
- 12.Forslund, O., B. G. Hansson, P. Rymark, and B. Bjerre. 1993. Human papillomavirus DNA in urine samples compared with that in simultaneously collected urethra and cervix samples. J. Clin. Microbiol. 31:1975-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forslund, O., B. G. Hansson, and B. Bjerre. 1994. Typing of human papillomaviruses by consensus polymerase chain reaction and a non-radioactive reverse dot blot hybridization. J. Virol. Methods 49:129-139. [DOI] [PubMed] [Google Scholar]
- 14.Gravitt, P. E., C. L. Peyton, T. Q. Alessi, C. M. Wheeler, F. Coutlee, A. Hildesheim, M. H. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho, G. Y. F., R. D. Burk, S. Klein, A. S. Kadish, C. J. Chang, P. Palan, J. Basu, R. Tachezy, R. Lewis, and S. Romney. 1995. Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J. Natl. Cancer Inst. 87:1365-1371. [DOI] [PubMed] [Google Scholar]
- 16.Ho, G. Y. F., R. Bierman, L. Beardsley, C. J. Chang, and R. D. Burk. 1998. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 338:423-428. [DOI] [PubMed] [Google Scholar]
- 17.. Houfflin Debarge, V., P. Collinet, D. Vinatier, A. Ego, A. Dewilde, F. Boman, and J. L. Leroy. 2003. Value of human papillomavirus testing after conization by loop electrosurgical excision for high-grade squamous intraepithelial lesions. Gynecol. Oncol. 90:587-592. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, M. V., P. J. F. Snijders, J. C. van den Brule, T. J. M. Helmerhorst, C. J. L. M. Meijer, and J. M. M. Walboomers. 1997. A general primer GP5+/GP6+-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs, M. V., J. M. M. Walboomers, P. J. F. Snijders, F. J. Voorhorst, R. H. M. Verheijen, N. Fransen-Daalmeijer, and C. J. L. M. Meijer. 2000. Distribution of 37 mucosotropic HPV types in women with cytologically normal cervical smears: the age-related patterns for high-risk and low-risk types. Int. J. Cancer 87:221-227. [PubMed] [Google Scholar]
- 20.Koss, L. G. (ed). 1979. Diagnostic cytology and its histologic basis, 3rd ed. Lippincott, Philadelphia, Pa.
- 21.Koutsky, L. A., K. K. Holmes, C. W. Critchlow, C. E. Stevens, J. Paavonen, A. M. Beckmann, T. A. DeRouen, D. A. Galloway, D. Vernon, and N. B. Kiviat. 1992. A cohort study of the risk of cervical intraepithelial neoplasia grade 2 or 3 in relation to papillomavirus infection. N. Engl. J. Med. 327:1272-1278. [DOI] [PubMed] [Google Scholar]
- 22.MacMahon, S., R. Peto, J. Cutler, R. Collins, P. Sorlie, J. Neaton, R. Abbott, J. Godwin, A. Dyer, and J. Stamler. 1990. Blood pressure, stroke and coronary heart disease. Part 1. Prolonged differences in blood pressure: Prospective observational studies corrected for the regression dilution bias. Lancet 335:765-774. [DOI] [PubMed] [Google Scholar]
- 23.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsauge, K. V. Shah, P. J. F. Snijders, C. J. L. M. Meijer, et al. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 24.Nobbenhuis, M. A. E., J. M. M. Walboomers, T. J. M. Helmerhorst, L. Rozendaal, A. J. Remmink, E. K. J. Risse, H. C. van der Linden, F. J. Voorhorst, P. Kenemans, and C. J. L. M. Meijer. 1999. Relation of human papillomavirus status to cervical lesions and consequences for cervical-cancer screening: a prospective study. Lancet 354:20-25. [DOI] [PubMed] [Google Scholar]
- 25.Nobbenhuis, M. A. E., T. J. M. Helmerhorst, A. J. C. van den Brule, L. Rozendaal, F. J. Voorhorst, P. D. Bezemer, R. H. M. Verheijen, and C. J. L. M. Meijer. 2001. Cytological regression and clearance of high-risk human papillomavirus in women with an abnormal cervical smear. Lancet 358:1782-1783. [DOI] [PubMed] [Google Scholar]
- 26.Nobbenhuis, M. A. E., C. J. L. M. Meijer, A. J. C. van den Brule, L. Rozendaal, F. J. Voorhorst, E. K. J. Rissa, R. H. M. Verheijen, and T. J. M. Helmerhorst. 2001. Addition of high-risk HPV testing improves the current guidelines on follow-up after treatment for cervical intraepithelial neoplasia. Br. J. Cancer 84:796-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paraskevaidis, E., M. Arbyn, A. Sotiriadis, E. Diakomanolis, P. Martin-Hirsch, G. Koliopoulos, G. Makrydimas, J. Tofoski, and D. H. Roukos. 2004. The role of HPV DNA testing in the follow-up period after treatment for CIN: a systematic review of the literature. Cancer Treat. Rev. 30:205-211. [DOI] [PubMed] [Google Scholar]
- 28.Parkin, M., P. Pisani, and J. Ferlay. 1999. Estimates of the worldwide incidence of 25 major cancers in 1990. Int. J. Cancer 80:827-841. [DOI] [PubMed] [Google Scholar]
- 29.Petry, K. U., S. Menton, M. Menton, F. van Loenen-Frosch, H. de Carvalho Gomes, B. Holz, B. Schopp, S. Garbrecht-Buettner, P. Davies, G. Boehmer, E. van den Akker, and T. Iftner. 2003. Inclusion of HPV testing in routine cervical cancer screening for women above 29 years in Germany: results for 8466 patients. Br. J. Cancer 88:1570-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peyton, C. L., M. Schiffman, A. T. Lörincz, W. C. Hunt, I. Mielzynska, C. Bratti, S. Eaton, A. Hildesheim, L. A. Morera, A. C. Rodriguez, R. Herrero, M. E. Sherman, and C. M. Wheeler. 1998. Comparison of PCR- and Hybrid Capture-based human papillomavirus detection systems using multiple cervical specimen collection strategies. J. Clin. Microbiol. 36:3248-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poljak, M., A. Brencic, K. Seme, A. Vince, and I. J. Marin. 1999. Comparative evaluation of first- and second-generation Digene Hybrid Capture assays for detection of human papillomaviruses associated with high or intermediate risk for cervical cancer. J. Clin. Microbiol. 37:796-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romney, S. L., G. Y. F. Ho, P. R. Palan, J. Basu, A. S. Kadish, S. Klein, M. Mikhail, R. J. Hagan, C. J. Chang, and R. D. Burk. 1997. Effects of β-carotene and other factors on outcome of cervical dysplasia and human papillomavirus infection. Gynecol. Oncol. 65:483-492. [DOI] [PubMed] [Google Scholar]
- 33.Rozendaal, L., J. M. M. Walboomers, J. C. van der Linden, F. J. Voorhorst, P. Kenemans, T. J. M. Helmerhorst, M. van Ballegooijen, and C. J. L. M. Meijer. 1996. PCR-based high-risk HPV test in cervical cancer screening gives objective risk assessment of women with cytomorphologically normal cervical smears. Int. J. Cancer 68:766-769. [DOI] [PubMed] [Google Scholar]
- 34.Schiffman, M. H., H. M. Bauer, R. N. Hoover, A. G. Glass, D. M. Cadell, B. B. Rush, D. R. Scott, M. E. Sherman, R. J. Kurman, S. Wacholder, C. K. Stanton, and M. M. Manos. 1993. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 85:958-964. [DOI] [PubMed] [Google Scholar]
- 35.Schneede, P., P. Hillemanns, F. Ziller, A. Hofstetter, E. Stockfleth, R. Arndt, and T. Meyer. 2001. Evaluation of HPV testing by Hybrid Capture II for routine gynaecologic screening. Acta Obstet. Gynecol. Scand. 80:750-752. [DOI] [PubMed] [Google Scholar]
- 36.Sherman, M. E., A. T. Lorincz, D. R. Scott, S. Wacholder, P. E. Castle, A. G. Glass, I. Mielzynska-Lohnas, B. B. Rush, and M. Schiffman. 2003. Baseline cytology, human papillomavirus testing, and risk for cervical neoplasia: a 10-year cohort analysis. J. Natl. Cancer Inst. 95:46-52. [DOI] [PubMed] [Google Scholar]
- 37.Venturoli, S., M. Cricca, F. Bonvicini, F. Giosa, F. R. Pulvirenti, C. Galli, M. Musiani, and M. Zerbini. 2002. Human papillomavirus DNA testing by PCR-ELISA and hybrid capture II from a single cytological specimen: concordance and correlation with cytological results. J. Clin. Virol. 25:177-185. [DOI] [PubMed] [Google Scholar]
- 38.Vince, A., N. Kutela, J. Iscic-Bes, V. Harni, M. Ivanisevic, Z. Sonicki, Z. Culig, and M. Poljak. 2002. Clinical utility of molecular detection of human papillomavirus in cervical samples by hybrid capture technology. J. Clin. Virol. 25:109-112. [DOI] [PubMed] [Google Scholar]
- 39.Walboomers, J. M. M., and C. J. L. M. Meijer. 1997. Do HPV-negative cervical carcinomas exist? J. Pathol. 181:253-254. [DOI] [PubMed] [Google Scholar]
- 40.Walboomers, J. M. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. F. Snijders, J. Peto, C. J. L. M. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]