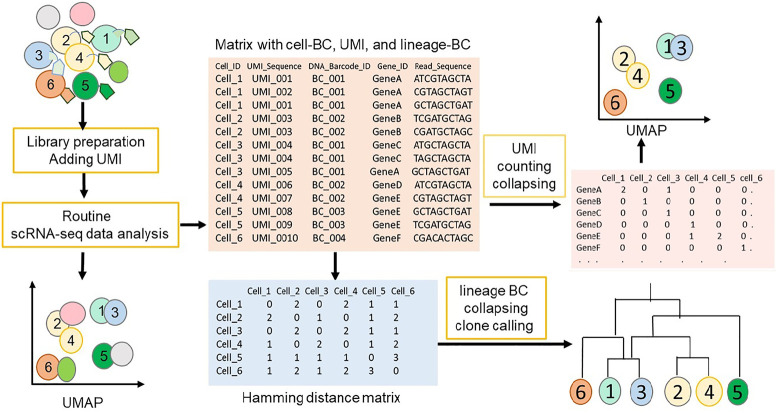
Figure 4.
Overall steps in scLT data analysis and barcode calling. Before scRNA-seq library preparation, only cells 1–6 were labeled with lineage barcodes. scLT data can first be analyzed using routine scRNA-seq data analysis steps, such as read alignment, UMI counting and collapsing, filtering, normalization, and clustering, including cells with and without lineage barcodes. Then, the data can be filtered based on UMI counts and lineage barcode availability to obtain cells with cell barcodes, UMIs, and lineage barcodes. This filtered data can be further analyzed using regular scRNA-seq steps or used for lineage analysis to construct lineage trees based on cell similarity determined by Hamming distance or other methods.