(
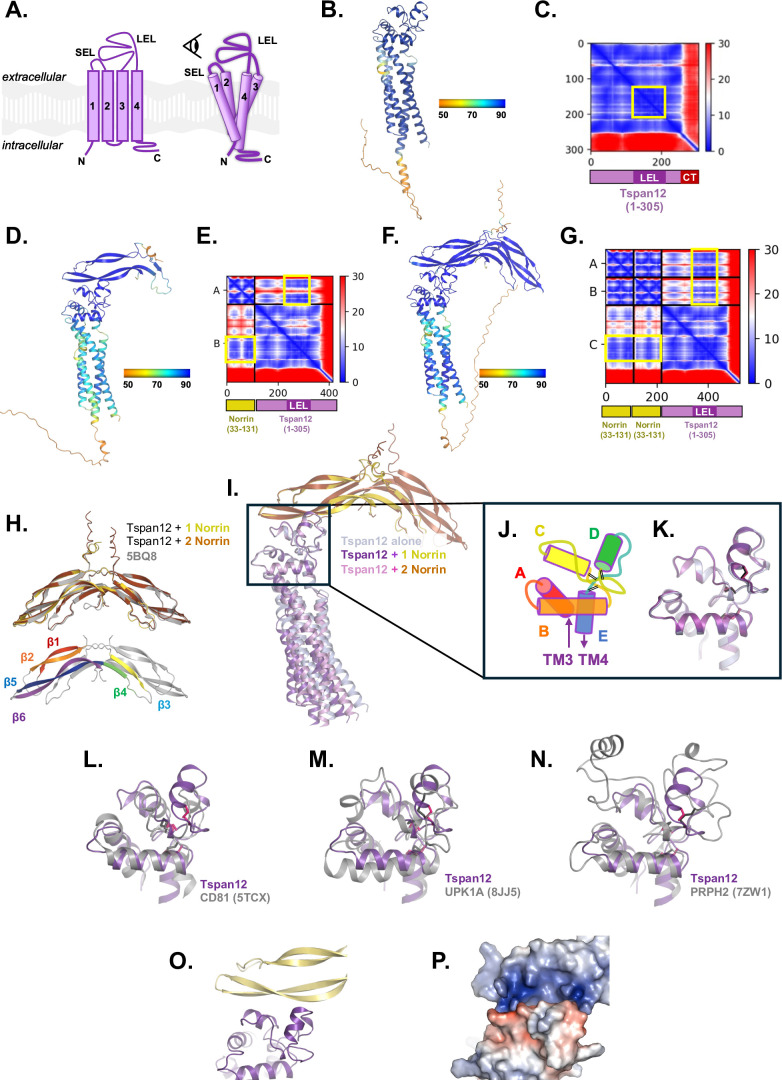
A) Cartoon of tetraspanin structure, comprised of transmembrane helices 1–4, a small extracellular loop (SEL, between helices 1 and 2) and a large extracellular loop (LEL, between helices 3 and 4). Eye icon indicates the viewing angle of AlphaFold models in the remainder of this figure (
B, D, F, I–P) relative to this cartoon. (
B) The structure of full-length human Tspan12 alone was predicted with AlphaFold. The best-scoring model is shown, colored by the per-residue predicted local distance difference test (pLDDT) confidence metric. (
C) The predicted aligned error (PAE) for the model in B. The position of the LEL (darker purple) appears as a darker blue square in the heat map with low PAE (yellow box). The position of C-terminal residues (‘CT‘, red) is indicated; the heat map shows high PAE values for the C-terminus relative to the rest of the protein, indicating poor prediction of the relative positioning of the C-terminus. (
D) The structure of full-length human Tspan12 together with one Norrin protomer (residues 25–133) was predicted with AlphaFold-Multimer and the best-scoring model is shown, colored by pLDDT. (
E) The predicted aligned error for the model shown in D. The position of Norrin, Tspan12, and the LEL along the axes are indicated. Note low PAE between the LEL and parts of Norrin (yellow boxes). (
F) The structure of full-length human Tspan12 together with two copies of Norrin (residues 25–133) was predicted with AlphaFold-Multimer and the best-scoring model is shown, colored by pLDDT. (
G) The predicted aligned error for the model shown in F. The position of both copies of Norrin, Tspan12, and the LEL along the axes are indicated. Note low PAE between the LEL and parts of both Norrin protomers (yellow boxes). (
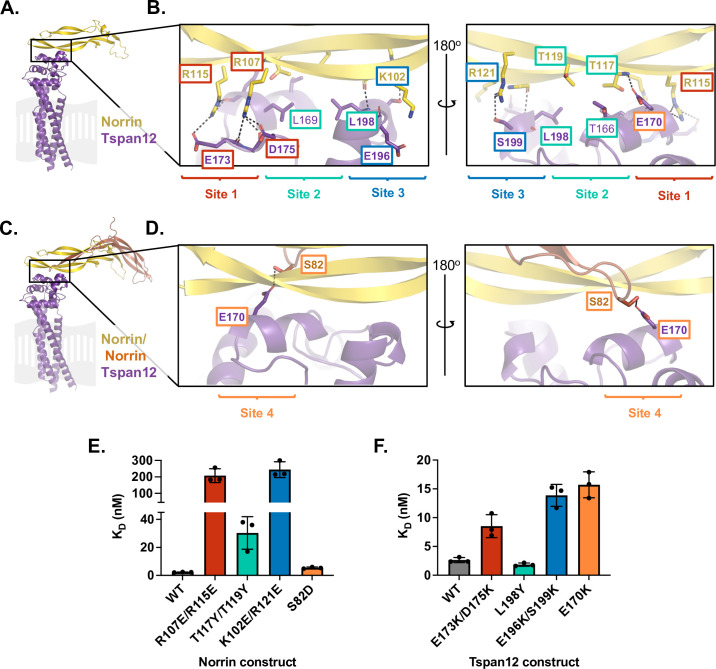
H) Top: The structure of Norrin within the predicted structure of Tspan12+1 Norrin protomer (yellow), and within the predicted structure of Tspan12+2 Norrin protomers (orange), matches the crystal structure of Norrin (5BQ8 chains A and B; gray) with RMSDs of 0.383 and 0.413 Å respectively. Below: β strands 1, 2, 3, 4, 5, and 6 of one Norrin protomer, colored red, orange, yellow, green, blue, and purple, respectively; strands 5 and 6 are predicted to comprise the Tspan12 binding site. (
I) The predicted model of Tspan12 alone (light blue) is similar to the predicted models with one (purple/yellow) or two (pink/orange) copies of Norrin, which each align with RMSDs of 1.247 and 1.057 Å, respectively, to the model of Tspan12 alone. Aligning only the LELs as shown gives RMSDs of 0.293 and 0.341 Å, respectively, and illustrates the slight variation in the predicted angle between the TMs and the LEL. The predicted position and orientation of Norrin relative to the LEL is unchanged between the one-protomer and two-protomer models. (
J) Tetraspanin LELs are composed of helices A, B, C, D, and E. Helices C and D are the least conserved and are implicated in binding partner interactions (
Susa et al., 2024). Black double lines represent disulfide bridges. Viewing angle is indicated by the eye in A. (
K) Close-up of aligned LELs from I. (
L) The predicted structure of the Tspan12 LEL (from the 1 Tspan12 : 1 Norrin protomer model; residues 123–215; purple) aligns to the experimentally determined structure of CD81 LEL (5TCX residues 123–198; gray), with an RMSD of 3.997 Å. CD81 is a C4 tetraspanin; disulfide bonds for Tspan12 and CD81 are shown in pink and black, respectively. (
M) The predicted Tspan12 LEL (purple) aligns to the Uroplakin 1A LEL (8JJ5, residues 125–226; gray), a C6 tetraspanin, with an RMSD of 4.215 Å. (
N) The predicted Tspan12 LEL (purple) aligns to the Peripherin-2 LEL (7ZW1, residues 126–258; gray), a C6 tetraspanin, with an RMSD of 2.528 Å. (
O) The predicted interaction between Norrin and Tspan12 involves helices C and D of the LEL and residues on β5 and β6 of Norrin. (
P) The predicted interaction between Norrin and Tspan12 colored by surface electrostatics (APBS) shows a highly polar interaction involving a basic patch on Norrin and an acidic patch on Tspan12.