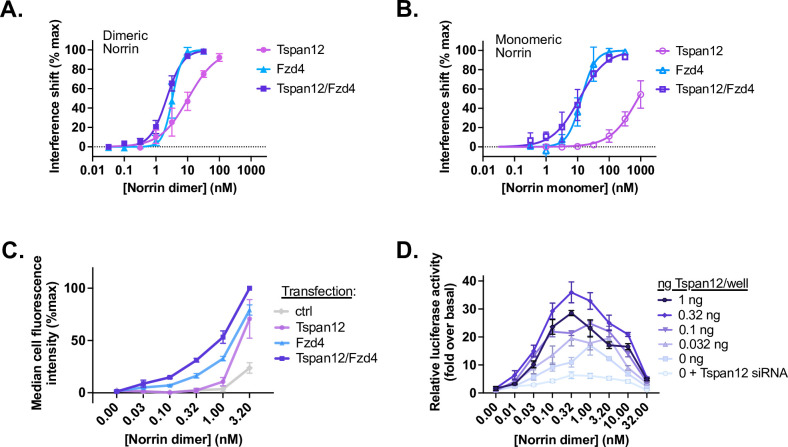
Figure 4. Tspan12 enhances Norrin-Fzd4 binding, cell-surface binding, and Norrin-stimulated β-catenin signaling at low Norrin concentrations.
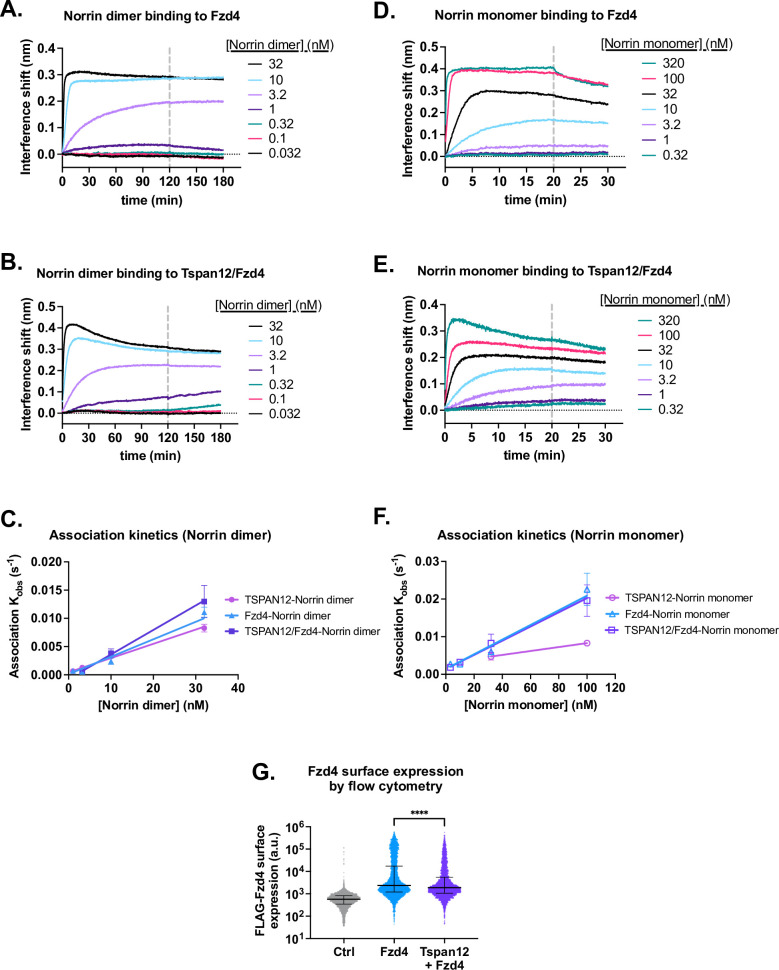
(A) Steady-state binding curves of monomeric Tspan12∆C, monomeric Fzd4, or heterodimeric Tspan12∆C/Fzd4∆C receptors in biotinylated nanodiscs binding to dimeric or (B) monomeric (C93A/C95A/C131A) Norrin by biolayer interferometry (BLI). Steady-state binding signal is plotted as a percent of Bmax for three independent replicates (mean ± SD). Affinities and kinetic constants are reported in Supplementary file 1. (C) Indicated concentrations of Norrin-1D4 dimer binding to Expi293 cells transfected with Fzd4, Tspan12, or both Fzd4 and Tspan12, detected with fluorescently labeled Rho1D4 antibody and quantified by flow cytometry. Mean ± SD of three independent experiments are plotted. Co-transfection of Tspan12 increased Norrin recruitment to Fzd4-transfected cells at 0.1, 0.32, 1, and 3.2 nM Norrin (two-tailed t-test p-values of 0.00026, 0.00079, 0.0049, and 0.0018, respectively). (D) β-Catenin pathway activation resulting from increasing concentrations of Norrin was assessed in Fzd1/2/4/5/7/8-knockout HEK293T cells transfected with Tspan12 siRNA or increasing amounts of Tspan12 plasmid, along with Fzd4 and TopFlash luciferase reporter plasmids. Data are plotted as mean ± SD from triplicate wells are representative of three independent experiments.