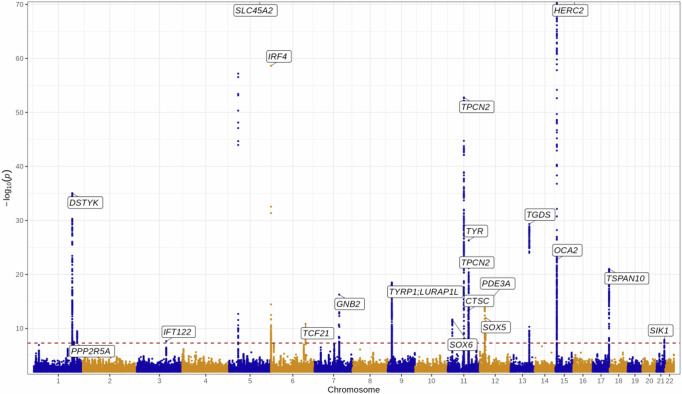
Fig. 3. Manhattan plot of GWAS results from the discovery cohort (UKBiobank, n = 37067).
The Y-axis represents the two-sided p-values from the linear mixed effects model. Lead variants identified by GCTA-COJO are annotated with the nearest gene. Points are truncated at −log10(p) = 70 for clarity. The dashed red line indicates genome-wide significance (p = 5 × 10−8) which is adjusted for multiple comparisons and the p-values are two-sided and calculated with the z-statistic. Source data are provided as a Source Data file.