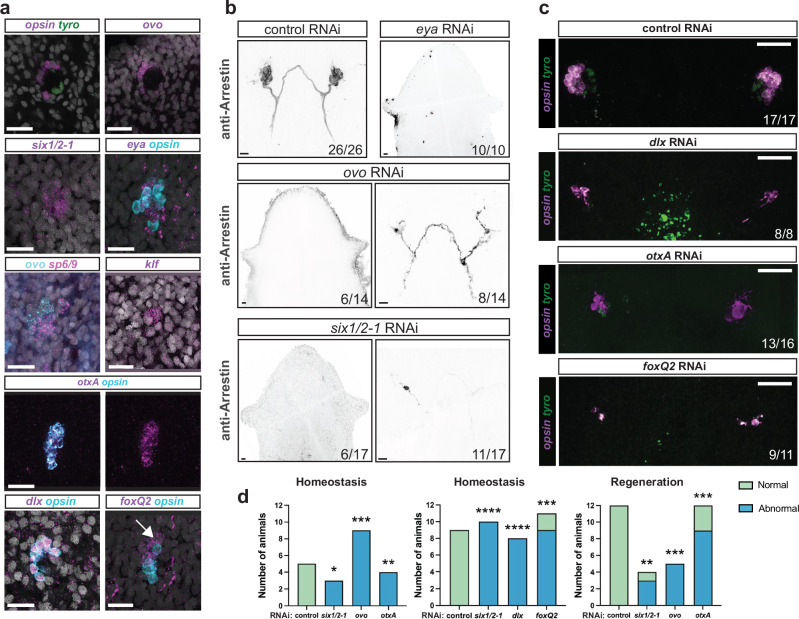
Fig. 4. The developmental program of planarian eye differentiation is largely conserved in cave planarians.
a Expression of key eye transcription factors in the intact cave planarian G. multidiverticulata, non-discernible eye morphotype. dlx+ and opsin+ double-positive signal shows colocalization in photoreceptor cells and not in the optic cup. foxQ2 and opsin show colocalization, but foxQ2 expression also occurs in a more anterior region near the photoreceptor cells (arrow). Representative images from n = 5 animals. b RNAi experiment phenotypes presenting normal (control), abnormal (ovo, six1/2-1 right panels), or absent (eya, ovo, six1/2-1 left panels) eyes. In control RNAi animals, 26 out of 26 presented normal eyes (26/26). Normal eyes contained photoreceptor neurons and photoreceptor axons visualized with the anti-Arrestin (VC-1) antibody. Six out of 14 ovo RNAi animals displayed no eye formation (6/14), and eight out of 14 displayed an abnormal eye (8/14), containing few photoreceptor cells and disorganized axon projections. Similarly, some treated animals with six1/2-1 RNAi displayed no eye formation (6/17), but the majority displayed abnormal eyes (11/17). c Abnormal phenotypes after eye transcription factor RNAi. Eye images shown of each experimental condition: Control (n = 26), six1/2-1 (n = 17), dlx (n = 8), ovo (n = 22), otxA (n = 16), and foxQ2 (n = 11). d RNAi during regeneration and homeostasis were blindly scored by two independent scorers, who classified the images as either normal (green) or abnormal (blue). The results were then compared using a Two-sided Fisher’s exact test, **p < 0.01, ***p < 0.001, ****p < 0.0001. RNAi experiments were performed with the G. multidiverticulata non-discernible eye morphotype.