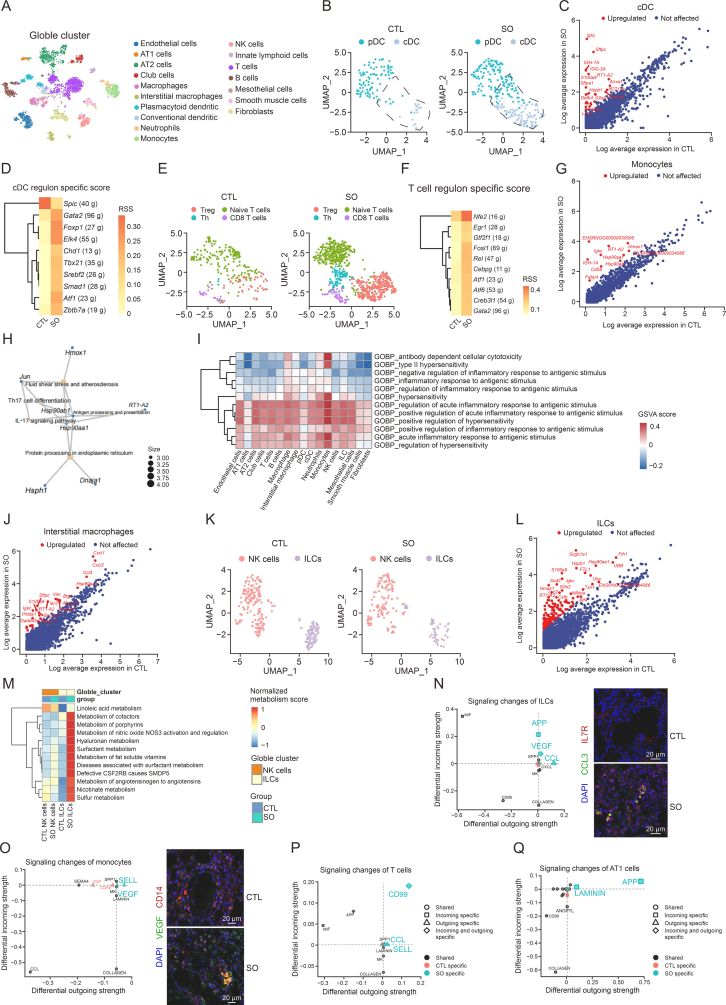
Fig. 1.
Single-cell RNA sequencing (scRNA-seq) was used to identify diverse cellular change in the lungs of the water extract of Siegesbeckia orientalis L. (SO)-treated rats. (A) t-distributed Stochastic neighbor embedding (t-SNE) plot showing the lung cells cluster from control (CTL) and SO groups. (B) Uniform manifold approximation and projection (UMAP) plot showing the dendritic cells (DCs) integration from two groups. (C) Conventional dendritic cells (cDCs) differentially expression genes (DEGs) analysis between CTL and SO groups. (D) cDCs regulon specific score reveals the differential regulon activation between CTL and SO. (E) Unsupervised clustering of T cells. (F) T cells regulon specific score showing the differential regulon activation between CTL and SO groups. (G) Monocytes DEGs analysis between CTL and SO groups. (H) Monocyte Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis based on the SO-upregulated DEGs. (I) Gene set variation analysis (GSVA) heatmap illustrating the expression of the inflammatory-related biological processes in all cell types within the SO group. (J) Interstitial macrophages DEGs analysis between CTL and SO groups. (K) UMAP plot showing innate lymphoid cells (ILCs) and natural killer (NK) cells from two groups. DEGs related to NK cells and ILCs based on the integrated dataset. (L) ILCs DEGs analysis between CTL and SO groups. (M) Metabolism heatmap to compare the metabolic changes in T cells and B cells between the CTL and SO groups. (N, O) Differential outgoing or incoming signaling pathways of ILCs and monocytes between CTL and SO groups from cell communication analysis. The expression of C−C motif chemokine ligand 3 (CCL3) in ILCs (N) and the expression of vascular endothelial growth factor (VEGF) in monocytes (O) were validated by immunofluorescence staining, respectively. (P, Q) Differential outgoing or incoming signaling pathways of T cells (P) and AT1 cells (Q) between CTL and SO groups from cell communication analysis. In Figs. 1C, G, J, and L, red dots represent the upregulated genes in SO when compared with CTL group and blue dots represent those not altered genes in both CTL and SO groups. pDC: plasmacytoid dendritic cells; RSS: regulon specific score; Th: T helper cell; IL-17: interleukin-17; NOS3: nitric oxide synthase 3; CSF2RB: colony stimulating factor 2 receptor subunit beta; SMDP5: pulmonary surfactant metabolism dysfunction-5; APP: amyloid precursor protein; SPP1: secreted phosphoprotein 1; CXCL: chemokine (C−X−C motif) ligand; MK: midkine; DAPI: 4’,6-diamidino-2’-phenylindole; IL-7R: IL-7 receptor; SEMA4: semaphorin 4; CSF: colony-stimulating factor; MIF: macrophage migration inhibitory factor; ANGPTL: angiopoietin like.