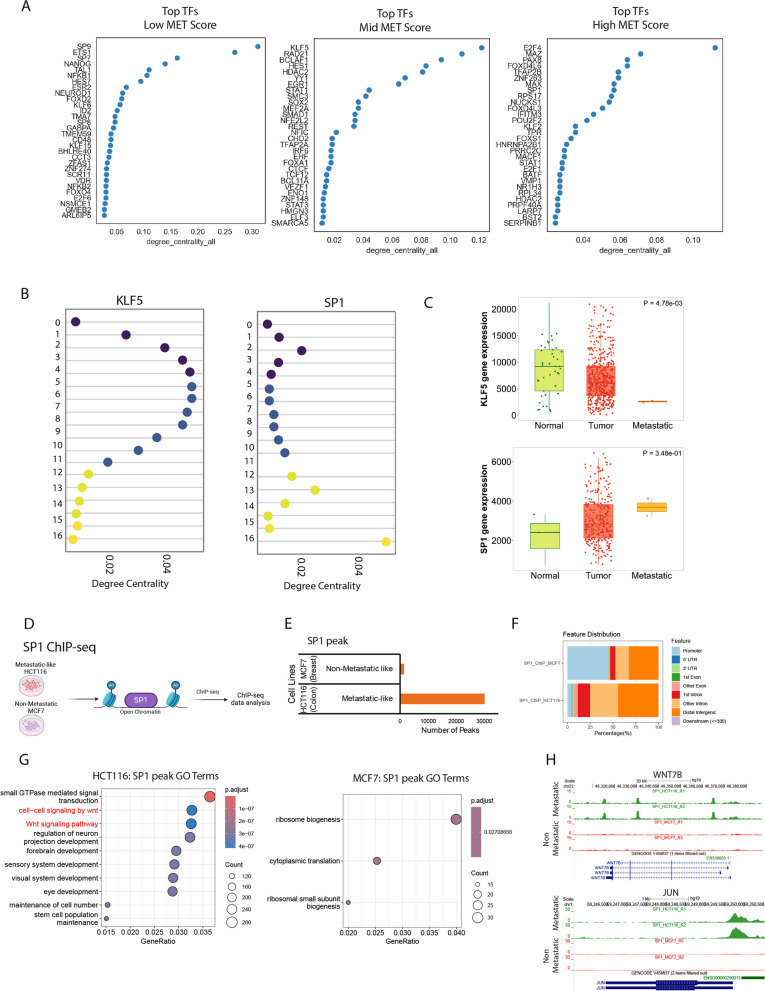
Fig. 7.
Reconstructing low to high metastatic regulatory networks conserved across cancers. A Transcription factors (TFs) within the gene regulatory network (GRN) associated with different metastatic stages. B Network dynamics of TFs across metastatic stages, coloured by MET_Score stage. C Expression levels of KLF5 and SP1 in normal, tumour, and metastatic samples, as measured by RNA sequencing (RNA-seq). D Genome browser tracks of SP1 ChIP-seq data, highlighting WNT target genes bound by SP1. D Schematic overview of the SP1 ChIP-seq data analysis. The SP1 ChIP-seq data were derived from ENCODE database for metastatic-like HCT116 and non-metastatic-like MCF7 cells and analysed using standard ChIP-seq analysis pipeline. E The barplot shows number of peaks detected for SP1 bound regions in HCT116 and MCF7 cells. F The barplot shows annotation of SP1 bound genes at promoter and non-promoter regions of the genome. G The dotplot represents top enriched pathways of SP1 bound genes in HCT116 and MCF7 cells. The top enriched pathways were derived using hallmark gene signatures from Molecular Signatures Database (MSigDB). H) The browser tracks show SP1 binding signal at hallmark WNT pathway genes