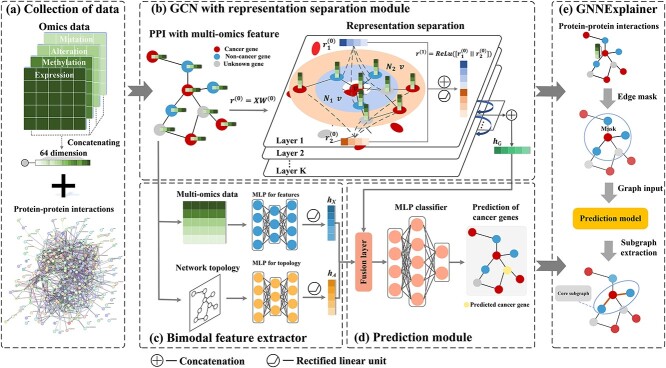
Figure 1.
Overview of SGCD. (a) Collection of PPIs and 64-dimensional multi-omics data. (b) GCN with RS Module. We leverage a GCN with RS to learn node embeddings from multi-omics and PPI networks. (c) Bimodal feature extractor. It incorporates two Multilayer Perceptrons (MLP) to separately embed the topological information and the multi-omics. (d) Prediction module. We combine the convolution-derived representation with the bimodal MLP-derived representation through a linear layer to estimate whether a gene serves as a driver gene. (e) GNNExplainer is adopted to identify cancer gene modules.