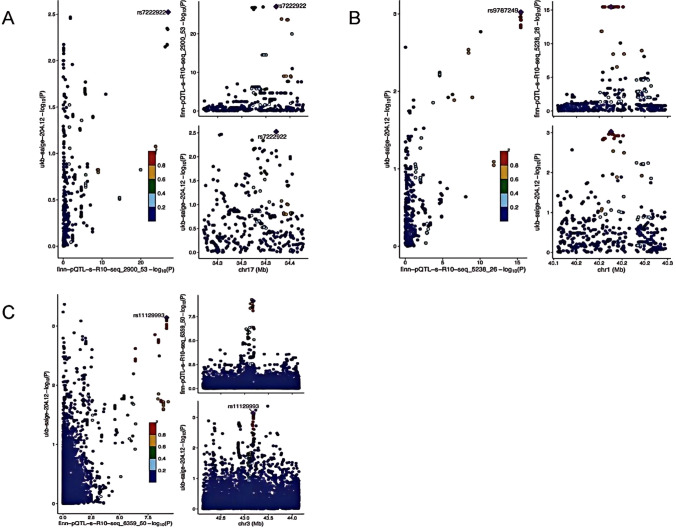
Fig. 4.
Genetic colocalization of CLL (A) CCL14 (B) PPIE (C) POGNT2. In this view, each dot is a genetic variant. Te SNP with the most notable P value with CLL is marked, and the colors of other SNPs depends on the digit size ordering of linkage disequilibrium (r2). SNPs with missing linkage disequilibrium information are also coded dark blue. In the LocusZoom plots, -log10 (P.gwas) for links with CLL risk are on the x-axes, and -log10 (P.pqtl) for relationship with the protein levels on the y-axes