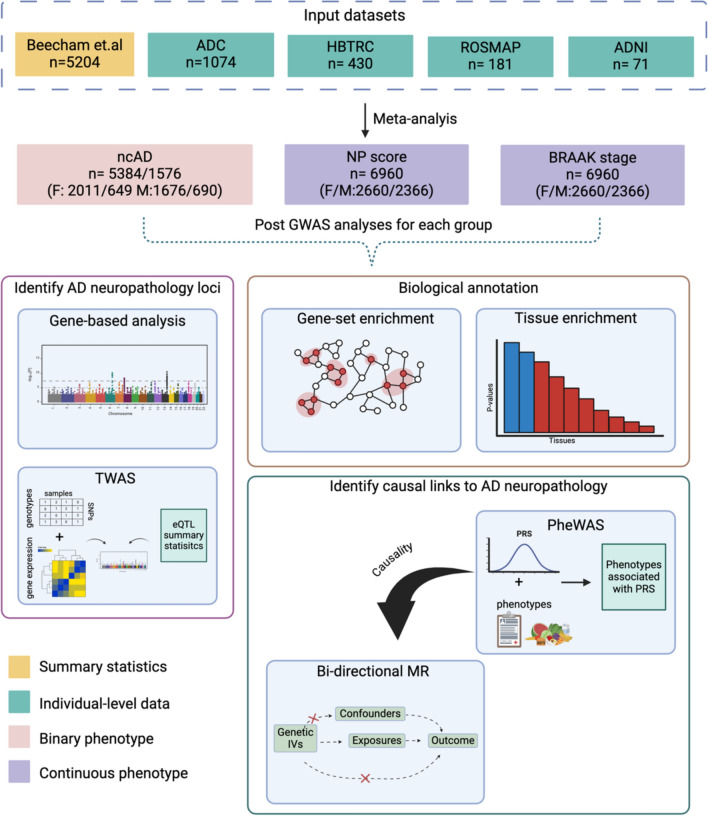
Fig. 1.
illustrates the comprehensive workflow of our study, outlining each key step from data integration to final analyses. We analyzed a total of 6,960 individuals by integrating data from 14 large-scale genetic, clinical, and neuropathology datasets, expanding upon previous Alzheimer’s Disease Genetics Consortium (ADGC) studies (Supplementary Table S1). These datasets included neuropathology assessments from the Alzheimer’s Disease Center (ADC), Religious Orders Study and Memory and Aging Project (ROSMAP), The Harvard Brain Tissue Resource Center (HBTRC), and the Alzheimer’s Disease Neuroimaging Initiative (ADNI), contributing 1756 additional samples. We conducted genome-wide association studies (GWAS) on three phenotypes: neuropathology-confirmed Alzheimer’s disease (ncAD), Braak stage, and NP score, applying stringent quality control procedures for both genotypes and variants. We performed sex-specific GWAS meta-analyses on 2660 females and 2366 males and conducted post-GWAS analyses using gene-based and gene-set approaches with FUMA and MAGMA, examining tissue specificity and gene set enrichment. Furthermore, transcriptome-wide association studies (TWAS) were performed across 13 GTEx v8 brain tissues, using the Joint-Tissue Imputation (JTI) method to identify gene regulation patterns. In addition, we conducted a PheWAS-based analysis using polygenic risk scores (PRS) to explore associations with 2248 UK Biobank phenotypes and performed Mendelian Randomization (MR) analysis to assess causal relationships between neuropathology traits and blood biomarkers from UK Biobank. Replication of significant findings was carried out using independent datasets. *ncAD: neuropathology-confirmed AD