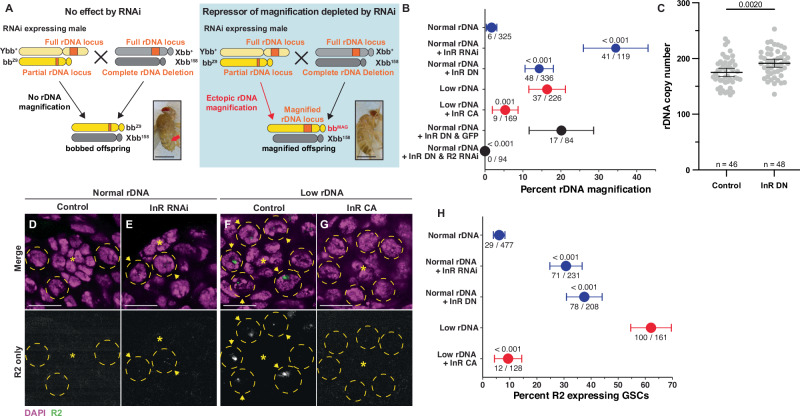
Fig. 2. InR represses rDNA magnification and R2 expression.
A Schematic to rapidly assess the function of candidate rDNA magnification repressors. RNAi targeting a candidate gene is expressed in the early germline of males harboring the bbZ9 X chromosome with the Y chromosome containing a wild-type rDNA locus (Ybb+). Males are mated to females harboring an X chromosome completely lacking rDNA (Xbb158), and the resultant bbZ9/Xbb158 daughters are assessed for rDNA magnification based on reversion of the bobbed phenotype (indicated by red arrowhead). Scale bar = 1 mm. B Frequency of offspring with ectopic rDNA magnification determined by the presence of wild-type cuticle. p-value determined by two-sided chi-squared test compared to control condition (Normal rDNA for Normal rDNA + InR RNAi (p = 2.2 × 10−16) and normal rDNA + InR DN (p = 2.46 × 10−8); Low rDNA for Low rDNA + InR CA (p = 1.25 × 10−3); Normal rDNA + InR DN & GFP for Normal rDNA + InR DN and R2 RNAi (p = 1.49 × 10−5)). Data presented are the percent of animals exhibiting rDNA magnification, with source data listed below, and ±95% confidence interval (CI). C rDNA CN assessed by ddPCR in individual bbZ9/Xbb158 daughters of control of InR dominant-negative expressing males. p-value determined by two-tailed Welch’s t-test. The data presented is a mean value ± 95% CI. D–G R2 RNA FISH images in normal (D, E) or low rDNA CN testes (F, G). DAPI in magenta, R2 in green. Asterisk (*) indicates the hub (stem cell niche), GSCs in yellow dotted circle. R2 positive GSCs indicated by a yellow arrowhead. Scale bar = 10 µm. H Frequency of GSCs expressing R2. p-value determined by two-sided chi-squared test compared to the control condition (Normal rDNA for Normal rDNA + InR RNAi (p = 2.2 × 10−16) and Normal rDNA + InR DN (p = 2.2 × 10−16); Low rDNA for Low rDNA + InR CA (p = 2.2 × 10−16)). Data presented is the percent of total GSCs observed that express R2, with source data listed below, and ±95% CI. Genotypes: bbZ9/Ybb+;; nos-Gal4/+ (Normal rDNA, Control), bbZ9/Ybb+;; nos-Gal4/UAS-InR RNAiJF01482 (Normal rDNA + InR RNAi), bbZ9/Ybb+;; nos-Gal4/UAS-InRK1409A (Normal rDNA + InR DN, InR DN), bbZ9/Ybb0;; nos-Gal4/ + (Low rDNA), bbZ9/Ybb0;; nos-Gal4/UAS-InRK414P (low rDNA + InR CA), bbZ9/Ybb+; UAS-InRK1409A/+; nos-Gal4/UAS-R2 RNAi-1 (InR DN + R2 RNAi), bbZ9/Ybb+; UAS-InRK1409A/+; nos-Gal4/UAS-GFP (Normal rDNA + GFP). Source data are provided as a Source Data file.