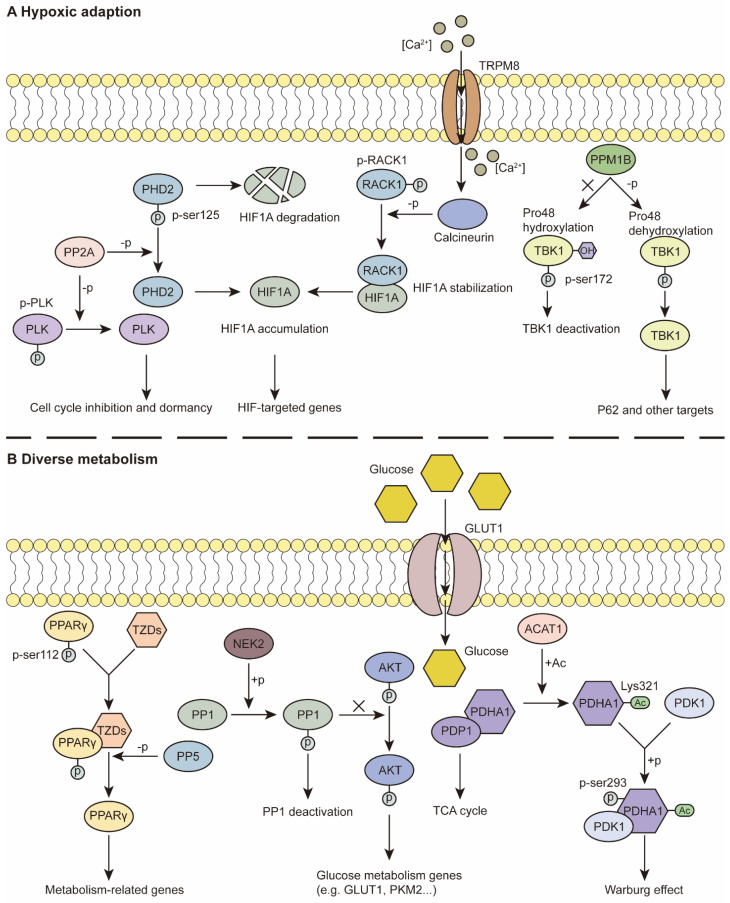
Figure 2.
PSPs in hypoxic adaption and diverse metabolism. (A) Hypoxic adaption: PP2A dephosphorylates PLK to inhibit cell cycle and promote dormancy. Dephosphorylation of PHD2 at Ser125 leaves HIF1A accumulation. The iron channel TRPM8, acts as a Ca2+ inflow channel, which facilitates the activation of calcineurin. Dephosphorylated RACK1 binds to HIF1A and stabilize HIF1A. Intact HIF1A regulates relative genes which contribute to hypoxic adaption. In normoxia, TBK1 is hydroxylated at Pro48, and can't be dephosphorylated by PPM1B. Under hypoxic conditions, dehydroxylated TBK1 can be dephosphorylated at Ser172, which leads to activation of downstream factors. (B) Diverse metabolism: TZDs can bind to PPARγ, which recruits PP5 to dephosphorylate PPARγ at Ser112. Dephosphorylated PPARγ is activated and targets various metabolism-related genes. PP1 is phosphorylated by NEK2. PP1 deactivation facilitates AKT regulation of glucose metabolism genes. PDP1/PDHA1 complex contributes to TCA cycle. Once acetylated on Lys321 by ACAT1, PDHA1 dissociates from PDP1 and binds to PDK1, which leads to Ser293 phosphorylation and the subsequent Warburg effect.