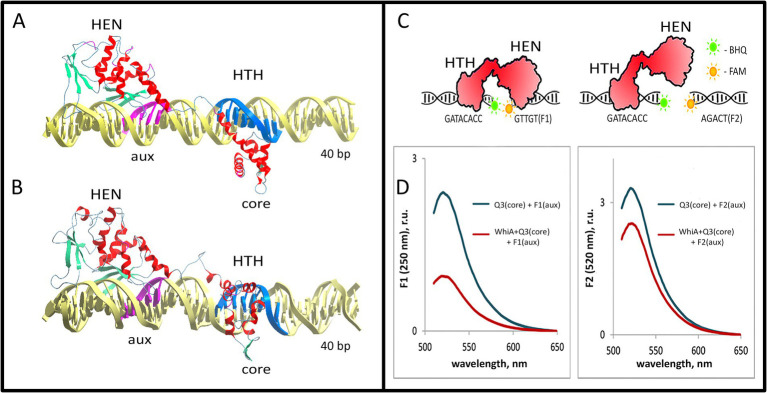
Figure 5.
Model of the Mgal-WhiA complex with the rpsJ operon promoter. (A,B) The structure of the Mgal-WhiA complex with DNA at the beginning (A) and at the end (B) of the molecular dynamics simulation. The complex (B) demonstrates DNA duplex distortion during the MD simulation. (C,D) The validation of the complex structure using the FRET experiment. (C) The schematic representation of the FRET experiment. The binding site of Mgal-WhiA was split into two halves containing either core (Q3 oligonucleotide, BHQ1-labeled) or aux motif (F1 oligonucleotide, 6-FAM-labeled). The control oligonucleotide featured mutated aux motif (F2 oligonucleotide, 6-FAM-labeled). (D) The assembly of the complete complex (WhiA+core+aux) resulted in significant quenching of the 6-FAM fluorescence in comparison to the control (WhiA+core+control). Thus Mgal-WhiA specifically recognizes aux motif and binds DNA in a bipartite mode.