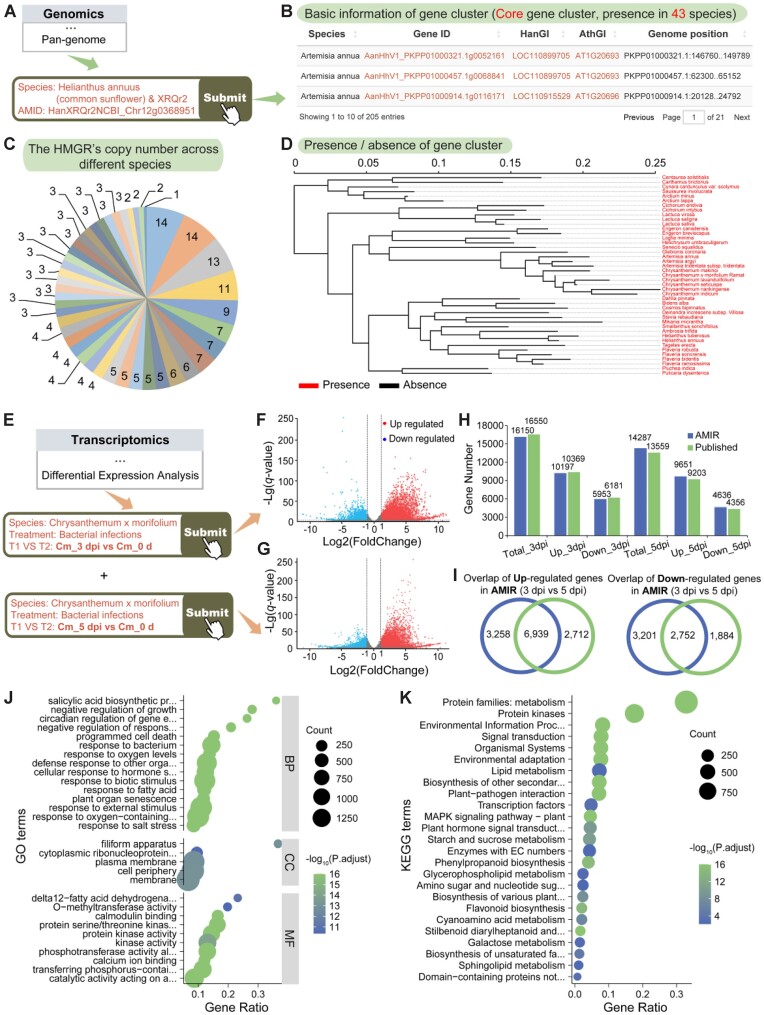
Figure 4.
Case study illustrating the utility of AMIR. (A) Process of searching for HMGR genes in the ‘Pan-genome’ module. (B) Pan-genome search results, including conservation of the queried gene, its distribution across different Asteraceae species and detailed gene information. (C) Number of homologous genes in each species. (D) Presence or absence of the queried gene in Asteraceae species. (E) Process of searching for DEGs in the ‘Differential expression analysis’ module. (F) Volcano plot of DEGs in Chrysanthemum morifolium leaves after 3 and 5 days of Alternaria sp. infection. (G) Comparison of the number of DEGs in AMIR with those reported in the original study. (H) Continuously upregulated genes after 3 and 5 days of Alternaria sp. infection. (I) Continuously downregulated genes after 3 and 5 days of Alternaria sp. infection. (J) GO enrichment analysis of continuously upregulated genes after 3 and 5 days of Alternaria sp. infection. (K) KEGG enrichment analysis of continuously upregulated genes after 3 and 5 days of Alternaria sp. infection.