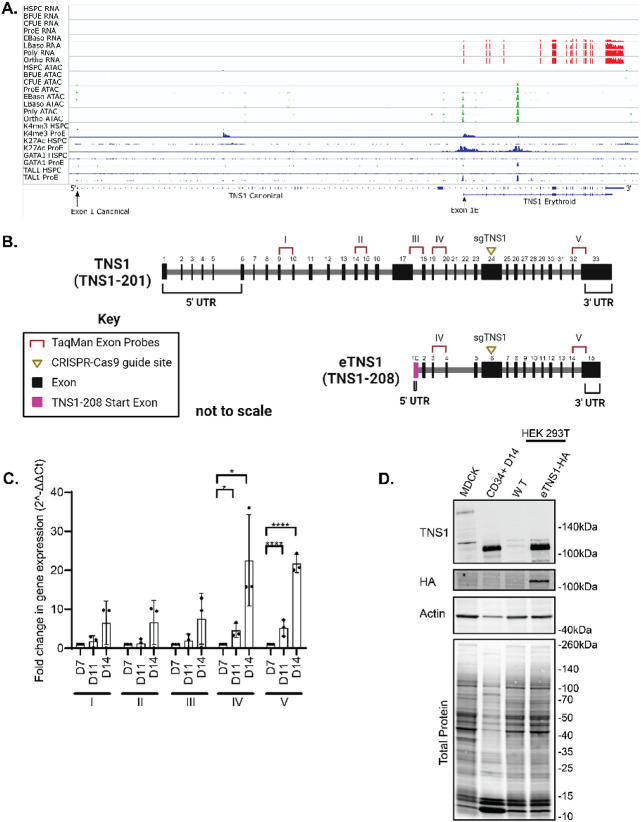
Figure 3. Human erythroblasts express a short isoform of TNS1 (eTNS1).
(A) mRNA expression of TNS1 exons during human erythroid differentiation from human stem and progenitor cells (HSPC) through terminal differentiation to orthrochromatic erythroblasts (orthoE). Exons encoding the canonical Ensembl TNS1 mRNA transcript and the erythroid TNS1 (eTNS1) mRNA transcript are shown at the bottom, with location of alternate exon 1s containing initiator methionines denoted by arrows. Top, red: mRNA expression, determined by RNA-seq, increases during terminal erythroid differentiation starting at the early basophilic erythroblast stage. Middle, green: Peaks of chromatin accessibility, determined by ATAC-seq, at the promoter and in a putative intron 4 enhancer increase during terminal erythroid differentiation. Bottom, blue. Peaks of histone marks and GATA1 and TAL1 transcription factor occupancy, determined by ChIP-seq, in HSPCa and proerythroblasts. (B) Comparison of canonical TNS1 gene to eTNS1 short isoform (not to scale), including the unique eTNS1 start exon (1E) highlighted in magenta. Pre-designed TaqMan primer-probe pairs (I-V) were chosen to span the canonical TNS1 cDNA at the specific exon sites shown (Key: TaqMan Exon Probes). sgRNA (yellow triangle) location designed for CRISPR-Cas9 knockout experiments. Schematic created with BioRender.com. (C) Quantification of mRNA expression (RT-qPCR) calculated as 2(−ΔΔC(T)) for all 5 probes on days 7, 11, and 14 of erythroid cultures. Fold change in gene expression normalized to average ΔCt value for day 7 cells for each of the 5 probes, using α-tubulin as a housekeeping gene. Values are mean ± SD from 3 individual erythroid cultures. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. (D) Representative western blot for eTNS1, HA, total actin and total protein in MDCK cells (full length TNS1), CD34+ day 14 cells (eTNS1), untransfected HEK293T cells and HEK293T cells transfected with an eTNS1-HA plasmid.