Figure 1. Whi3 condensate variability at hyphal tips and around nuclei is correlated with variability in hyphal elongation and cell cycle state.
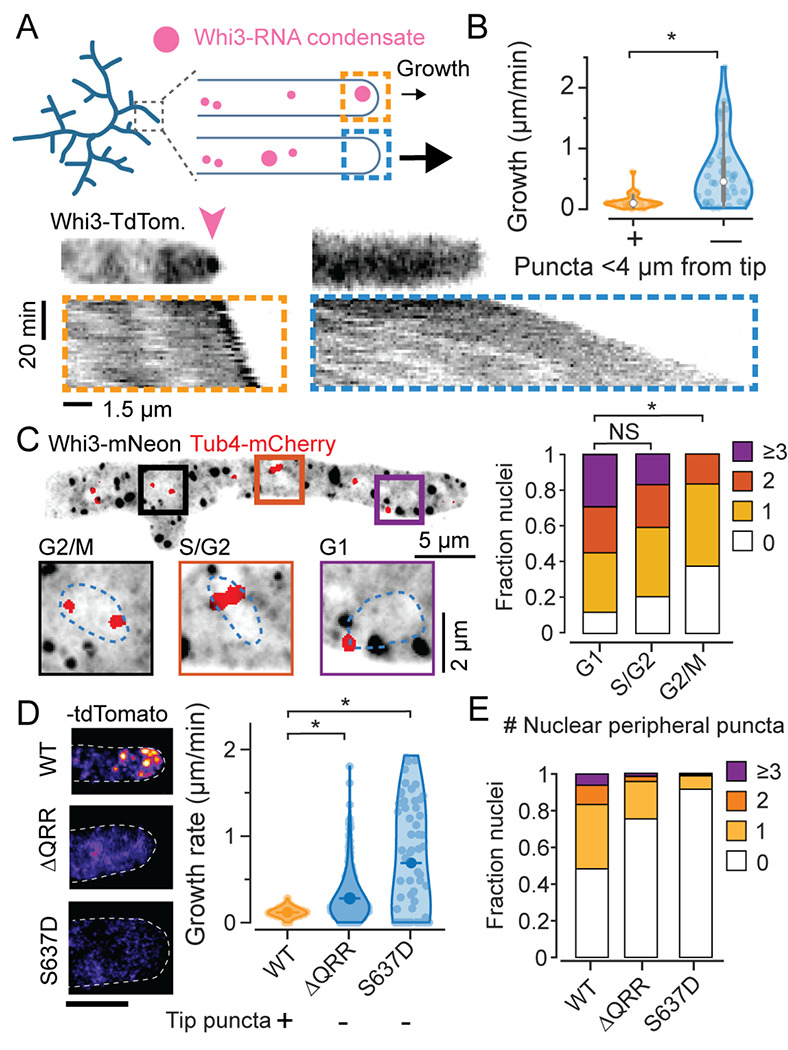
A. Schematic illustration of Whi3 condensates and tip elongation in Ashbya gossypii (top) and representative timelapse kymographs of hypha expressing Whi3:tdTomato (bottom). Kymographs were created from MIPs of 3D lightsheet timelapses. Orange box shows an example of a hypha with a prominent visible condensate at tip, marked by a pink arow, and the blue box shows an example of a hypha without a large tip-proximal condensate. Scale bar x: 1.5 μm, time: 20 min, origin on top left.
B. Instantaneous hyphal growth extracted from a total of 21 movies, split into 40 clips with or without tip proximal condensate (<4 μm from tip) each. * = p<0.01 using KS test.
C. Representative MIP of Tub4-mCherry expressing plasmidic Whi3-mNeon, scale bar 5 μm. Insets shows G1, S/G2 and G2/M nuclei with variable numbers of nuclear-peripheral Whi3 puncta, scale bar 2 μm. Stacked histograms quantifying relative nuclear proportions having varying peri-nuclear Whi3 puncta counts pooled from 3 biological replicates from a total of 238 G1, 88 S/G2, 24 G2/M phase nuclei. * = p<0.01 using KS test.
D. Representative images of WT Whi3:mNeon, non-condensate-forming whi3-QRR:tdTomato and whi3 S637D:tdTomato Ashbya hyphae (left) and hyphal elongation rates (right) measured using ConA pulse labeling. N>40 hyphae from >10 cells per strain. Scale bar 5 μm.
E. Stacked histograms quantifying relative nuclear proportions having varying peri-nuclear Whi3 puncta counts, from WT Whi3-tdTomato, n=180, whi3(ΔQRR):tdTomato, n=148, whi3(S637D):tdTomato, n=194.
